User info:
• About
• File formats
• Miller features in the context of the Unix toolkit
• Record-heterogeneity
• Performance
• Why call it Miller?
• How original is Miller?
• Reference
• Data examples
• Internationalization
• Things to do
Developer info:
• Compiling, portability, dependencies, and testing
• Why C?
• Contact information
• GitHub repo
• On-line help
• then-chaining
• I/O options
• Formats
• Record/field/pair separators
• Number formatting
• Data transformations
• cat
• check
• count-distinct
• cut
• filter
• group-by
• group-like
• having-fields
• head
• histogram
• join
• label
• put
• regularize
• rename
• reorder
• sort
• stats1
• stats2
• step
• tac
• tail
• top
• uniq
• Functions for filter and put
• Data types
• Null data
Command overview
Whereas the Unix toolkit is made of the separate executables cat, tail, cut, sort, etc., Miller has subcommands, invoked as follows:
mlr tac *.dat mlr cut --complement -f os_version *.dat mlr sort -f hostname,uptime *.dat
| Commands | Description |
|---|---|
| cat, cut, head, sort, tac, tail, top, uniq | Analogs of their Unix-toolkit namesakes, discussed below as well as in Miller features in the context of the Unix toolkit |
| filter, put, step | awk-like functionality |
| histogram, stats1, stats2 | Statistically oriented |
| group-by, group-like, having-fields | Particularly oriented toward Record-heterogeneity, although all Miller commands can handle heterogeneous records |
| count-distinct, label, rename, rename, reorder | These draw from other sources (see also How original is Miller?): count-distinct is SQL-ish, and rename can be done by sed (which does it faster: see Performance). |
On-line help
Examples:
$ mlr --help
Usage: mlr [I/O options] {verb} [verb-dependent options ...] {file names}
Verbs:
cat check count-distinct cut filter group-by group-like having-fields head
histogram join label put regularize rename reorder sort stats1 stats2 step tac tail top
uniq
Please use "mlr {verb name} --help" for verb-specific help.
Please use "mlr --help-all-verbs" for help on all verbs.
Functions for filter and put:
abs acos acosh asin asinh atan atan2 atanh cbrt ceil cos cosh erf erfc exp
expm1 floor invqnorm log log10 log1p max min pow qnorm round roundm sin sinh sqrt tan
tanh urand + - - * / % ** == != > >= < <= && || ! strlen sub tolower toupper .
boolean float int string hexfmt fmtnum gmt2sec sec2gmt systime
Please use "mlr --help-function {function name}" for function-specific help.
Please use "mlr --help-all-functions" or "mlr -f" for help on all functions.
Data-format options, for input, output, or both:
--dkvp --idkvp --odkvp Delimited key-value pairs, e.g "a=1,b=2" (default)
--nidx --inidx --onidx Implicitly-integer-indexed fields (Unix-toolkit style)
--csv --icsv --ocsv Comma-separated value (or tab-separated with --fs tab, etc.)
--pprint --ipprint --opprint --right Pretty-printed tabular (produces no output until all input is in)
--xtab --ixtab --oxtab Pretty-printed vertical-tabular
-p is a keystroke-saver for --nidx --fs space --repifs
Separator options, for input, output, or both:
--rs --irs --ors Record separators, defaulting to newline
--fs --ifs --ofs --repifs Field separators, defaulting to ","
--ps --ips --ops Pair separators, defaulting to "="
Notes (as of Miller v2.0.0):
* RS/FS/PS are used for DKVP, NIDX, and CSVLITE formats where they must be single-character.
* For CSV, PPRINT, and XTAB formats, RS/FS/PS command-line options are ignored.
* DKVP, NIDX, CSVLITE, PPRINT, and XTAB formats are intended to handle platform-native text data.
In particular, this means LF line-terminators on Linux/OSX.
* CSV is intended to handle RFC-4180-compliant data.
In particular, this means it *only* handles CRLF line-terminators.
* This will change in v2.1.0, at which point there will be a (default-off) LF-termination option
for CSV, multi-char RS/FS/PS, and double-quote support for DKVP.
Double-quoting for CSV:
--quote-all Wrap all fields in double quotes
--quote-none Do not wrap any fields in double quotes, even if they have OFS or ORS in them
--quote-minimal Wrap fields in double quotes only if they have OFS or ORS in them
--quote-numeric Wrap fields in double quotes only if they have numbers in them
Numerical formatting:
--ofmt {format} E.g. %.18lf, %.0lf. Please use sprintf-style codes for double-precision.
Applies to verbs which compute new values, e.g. put, stats1, stats2.
See also the fmtnum function within mlr put (mlr --help-all-functions).
Other options:
--seed {n} with n of the form 12345678 or 0xcafefeed. For put/filter urand().
Output of one verb may be chained as input to another using "then", e.g.
mlr stats1 -a min,mean,max -f flag,u,v -g color then sort -f color
Please see http://johnkerl.org/miller/doc and/or http://github.com/johnkerl/miller for more information.
This is Miller version >= v2.0.0.
$ mlr sort --help
Usage: mlr sort {flags}
Flags:
-f {comma-separated field names} Lexical ascending
-n {comma-separated field names} Numerical ascending; nulls sort last
-nf {comma-separated field names} Numerical ascending; nulls sort last
-r {comma-separated field names} Lexical descending
-nr {comma-separated field names} Numerical descending; nulls sort first
Sorts records primarily by the first specified field, secondarily by the second field, and so on.
Example:
mlr sort -f a,b -nr x,y,z
which is the same as:
mlr sort -f a -f b -nr x -nr y -nr z
then-chaining
In accord with the Unix philosophy, you can pipe data into or out of Miller. For example:mlr cut --complement -f os_version *.dat | mlr sort -f hostname,uptime
% cat piped.sh mlr cut -x -f i,y data/big | mlr sort -n y > /dev/null % time sh piped.sh real 0m2.828s user 0m3.183s sys 0m0.137s % cat chained.sh mlr cut -x -f i,y then sort -n y data/big > /dev/null % time sh chained.sh real 0m2.082s user 0m1.933s sys 0m0.137s
mlr cut --complement -f os_version then sort -f hostname,uptime *.dat
I/O options
Formats
Options:--dkvp --idkvp --odkvp --nidx --inidx --onidx --csv --icsv --ocsv --csvlite --icsvlite --ocsvlite --pprint --ipprint --ppprint --right --xtab --ixtab --oxtabThese are as discussed in File formats, with the exception of --right which makes pretty-printed output right-aligned:
$ mlr --opprint cat data/small a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 wye pan 5 0.5732889198020006 0.8636244699032729 |
$ mlr --opprint --right cat data/small a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 wye pan 5 0.5732889198020006 0.8636244699032729 |
-
Use --csv, --pprint, etc. when the input and output formats are the same.
Use --icsv --opprint, etc. when you want format conversion as part of what Miller does to your data.
DKVP (key-value-pair) format is the default for input and output. So,
--oxtab is the same as --idkvp --oxtab.
Record/field/pair separators
Miller has record separators IRS and ORS, field separators IFS and OFS, and pair separators IPS and OPS. For example, in the DKVP line a=1,b=2,c=3, the record separator is newline, field separator is comma, and pair separator is the equals sign. These are the default values. Options:--rs --irs --ors --fs --ifs --ofs --repifs --ps --ips --ops
-
You can change a separator from input to output via e.g. --ifs =
--ofs :. Or, you can specify that the same separator is to be used for
input and output via e.g. --fs :.
The pair separator is only relevant to DKVP format.
Pretty-print and xtab formats ignore the separator arguments altogether.
The --repifs means that multiple successive occurrences of the
field separator count as one. For example, in CSV data we often signify nulls
by empty strings, e.g. 2,9,,,,,6,5,4. On the other hand, if the field
separator is a space, it might be more natural to parse 2 4 5 the
same as 2 4 5: --repifs --ifs ' ' lets this happen. In fact,
the --ipprint option above is internally implemented in terms of
--repifs.
Just write out the desired separator, e.g. --ofs '|'. But you
may use the symbolic names newline, space, tab,
pipe, or semicolon if you like.
Number formatting
The command-line option --ofmt {format string} is the global number format for commands which generate numeric output, e.g. stats1, stats2, histogram, and step, as well as mlr put. Examples:--ofmt %.9le --ofmt %.6lf --ofmt %.0lf
$ echo 'x=3.1,y=4.3' | mlr put '$z=fmtnum($x*$y,"%08lf")' x=3.1,y=4.3,z=13.330000
$ echo 'x=0xffff,y=0xff' | mlr put '$z=fmtnum(int($x*$y),"%08llx")' x=0xffff,y=0xff,z=00feff01
$ echo 'x=0xffff,y=0xff' | mlr put '$z=hexfmt($x*$y)' x=0xffff,y=0xff,z=0xfeff01
Data transformations
cat
Most useful for format conversions (see File formats), and concatenating multiple same-schema CSV files to have the same header:
$ cat a.csv a,b,c 1,2,3 4,5,6 |
$ cat b.csv a,b,c 7,8,9 |
$ mlr --csv cat a.csv b.csv a,b,c 1,2,3 4,5,6 7,8,9 |
$ mlr --icsv --oxtab cat a.csv b.csv a 1 b 2 c 3 a 4 b 5 c 6 a 7 b 8 c 9 |
check
$ mlr check --help Usage: mlr check Consumes records without printing any output. Useful for doing a well-formatted check on input data.
count-distinct
$ mlr count-distinct --help
Usage: mlr count-distinct [options]
-f {a,b,c} Field names for distinct count.
Prints number of records having distinct values for specified field names. Same as uniq -c.
$ mlr count-distinct -f a,b then sort -nr count data/medium a=zee,b=wye,count=455 a=pan,b=eks,count=429 a=pan,b=pan,count=427 a=wye,b=hat,count=426 a=hat,b=wye,count=423 a=pan,b=hat,count=417 a=eks,b=hat,count=417 a=eks,b=eks,count=413 a=pan,b=zee,count=413 a=zee,b=hat,count=409 a=eks,b=wye,count=407 a=zee,b=zee,count=403 a=pan,b=wye,count=395 a=wye,b=pan,count=392 a=zee,b=eks,count=391 a=zee,b=pan,count=389 a=hat,b=eks,count=389 a=wye,b=eks,count=386 a=hat,b=zee,count=385 a=wye,b=zee,count=385 a=hat,b=hat,count=381 a=wye,b=wye,count=377 a=eks,b=pan,count=371 a=hat,b=pan,count=363 a=eks,b=zee,count=357
cut
$ mlr cut --help
Usage: mlr cut [options]
-f {a,b,c} Field names to include for cut.
-o Retain fields in the order specified here in the argument list.
Default is to retain them in the order found in the input data.
-x|--complement Exclude, rather that include, field names specified by -f.
Passes through input records with specified fields included/excluded.
$ mlr --opprint cat data/small a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 wye pan 5 0.5732889198020006 0.8636244699032729 |
$ mlr --opprint cut -f y,x,i data/small i x y 1 0.3467901443380824 0.7268028627434533 2 0.7586799647899636 0.5221511083334797 3 0.20460330576630303 0.33831852551664776 4 0.38139939387114097 0.13418874328430463 5 0.5732889198020006 0.8636244699032729 |
$ echo 'a=1,b=2,c=3' | mlr cut -f b,c,a a=1,b=2,c=3 |
$ echo 'a=1,b=2,c=3' | mlr cut -o -f b,c,a b=2,c=3,a=1 |
filter
$ mlr filter --help
Usage: mlr filter [-v] {expression}
Prints records for which {expression} evaluates to true.
With -v, first prints the AST (abstract syntax tree) for the expression, which
gives full transparency on the precedence and associativity rules of Miller's grammar.
Please use a dollar sign for field names and double-quotes for string literals.
Miller built-in variables are NF NR FNR FILENUM FILENAME PI E.
Examples:
mlr filter 'log10($count) > 4.0'
mlr filter 'FNR == 2 (second record in each file)'
mlr filter 'urand() < 0.001' (subsampling)
mlr filter '$color != "blue" && $value > 4.2'
mlr filter '($x<.5 && $y<.5) || ($x>.5 && $y>.5)'
Please see http://johnkerl.org/miller/doc/reference.html for more information including function list.
$ mlr filter 'FNR == 2' data/small* a=eks,b=pan,i=2,x=0.7586799647899636,y=0.5221511083334797 1=pan,2=pan,3=1,4=0.3467901443380824,5=0.7268028627434533 a=wye,b=eks,i=10000,x=0.734806020620654365,y=0.884788571337605134
$ mlr --opprint filter '$a == "pan" || $b == "wye"' data/small a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463
$ mlr --opprint filter '($x > 0.5 && $y > 0.5) || ($x < 0.5 && $y < 0.5)' then stats2 -a corr -f x,y data/medium x_y_corr 0.756439 |
$ mlr --opprint filter '($x > 0.5 && $y < 0.5) || ($x < 0.5 && $y > 0.5)' then stats2 -a corr -f x,y data/medium x_y_corr -0.747994 |
mlr --opprint filter ' ($x > 0.5 && $y < 0.5) || ($x < 0.5 && $y > 0.5)' \ then stats2 -a corr -f x,y data/medium
group-by
$ mlr group-by --help
Usage: mlr group-by {comma-separated field names}
Outputs records in batches having identical values at specified field names.
$ mlr --opprint group-by a data/small a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 eks wye 4 0.38139939387114097 0.13418874328430463 wye wye 3 0.20460330576630303 0.33831852551664776 wye pan 5 0.5732889198020006 0.8636244699032729 |
$ mlr --opprint sort -f a data/small a b i x y eks pan 2 0.7586799647899636 0.5221511083334797 eks wye 4 0.38139939387114097 0.13418874328430463 pan pan 1 0.3467901443380824 0.7268028627434533 wye wye 3 0.20460330576630303 0.33831852551664776 wye pan 5 0.5732889198020006 0.8636244699032729 |
group-like
$ mlr group-like --help Usage: mlr group-like Outputs records in batches having identical field names.
$ mlr cat data/het.dkvp resource=/path/to/file,loadsec=0.45,ok=true record_count=100,resource=/path/to/file resource=/path/to/second/file,loadsec=0.32,ok=true record_count=150,resource=/path/to/second/file resource=/some/other/path,loadsec=0.97,ok=false |
$ mlr --opprint group-like data/het.dkvp resource loadsec ok /path/to/file 0.45 true /path/to/second/file 0.32 true /some/other/path 0.97 false record_count resource 100 /path/to/file 150 /path/to/second/file |
having-fields
$ mlr having-fields --help
Usage: mlr having-fields [options]
--at-least {a,b,c}
--which-are {a,b,c}
--at-most {a,b,c}
Conditionally passes through records depending on each record's field names.
$ mlr cat data/het.dkvp resource=/path/to/file,loadsec=0.45,ok=true record_count=100,resource=/path/to/file resource=/path/to/second/file,loadsec=0.32,ok=true record_count=150,resource=/path/to/second/file resource=/some/other/path,loadsec=0.97,ok=false |
$ mlr having-fields --at-least resource data/het.dkvp resource=/path/to/file,loadsec=0.45,ok=true record_count=100,resource=/path/to/file resource=/path/to/second/file,loadsec=0.32,ok=true record_count=150,resource=/path/to/second/file resource=/some/other/path,loadsec=0.97,ok=false |
$ mlr having-fields --which-are resource,ok,loadsec data/het.dkvp resource=/path/to/file,loadsec=0.45,ok=true resource=/path/to/second/file,loadsec=0.32,ok=true resource=/some/other/path,loadsec=0.97,ok=false |
head
$ mlr head --help
Usage: mlr head [options]
-n {count} Head count to print; default 10
-g {a,b,c} Optional group-by-field names for head counts
Passes through the first n records, optionally by category.
$ mlr --opprint head -n 4 data/medium a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 |
$ mlr --opprint head -n 1 -g b data/medium a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 wye wye 3 0.20460330576630303 0.33831852551664776 eks zee 7 0.6117840605678454 0.1878849191181694 zee eks 17 0.29081949506712723 0.054478717073354166 wye hat 24 0.7286126830627567 0.19441962592638418 |
histogram
$ mlr histogram --help
Usage: mlr histogram [options]
-f {a,b,c} Value-field names for histogram counts
--lo {lo} Histogram low value
--hi {hi} Histogram high value
--nbins {n} Number of histogram bins
Just a histogram. Input values < lo or > hi are not counted.
$ mlr --opprint put '$x2=$x**2;$x3=$x2*$x' then histogram -f x,x2,x3 --lo 0 --hi 1 --nbins 10 data/medium bin_lo bin_hi x_count x2_count x3_count 0.000000 0.100000 1072 3231 4661 0.100000 0.200000 938 1254 1184 0.200000 0.300000 1037 988 845 0.300000 0.400000 988 832 676 0.400000 0.500000 950 774 576 0.500000 0.600000 1002 692 476 0.600000 0.700000 1007 591 438 0.700000 0.800000 1007 560 420 0.800000 0.900000 986 571 383 0.900000 1.000000 1013 507 341
join
$ mlr join --help
Usage: mlr join [options]
Joins records from specified left file name with records from all file names at the end of the Miller argument list.
Functionality is essentially the same as the system "join" command, but for record streams.
Options:
-f {left file name}
-j {a,b,c} Comma-separated join-field names for output
-l {a,b,c} Comma-separated join-field names for left input file; defaults to -j values if omitted.
-r {a,b,c} Comma-separated join-field names for right input file(s); defaults to -j values if omitted.
--lp {text} Additional prefix for non-join output field names from the left file
--rp {text} Additional prefix for non-join output field names from the right file(s)
--np Do not emit paired records
--ul Emit unpaired records from the left file
--ur Emit unpaired records from the right file(s)
-u Enable unsorted input. In this case, the entire left file will be loaded into memory.
Without -u, records must be sorted lexically by their join-field names, else not all
records will be paired.
File-format options default to those for the right file names on the Miller argument list, but may be overridden
for the left file as follows. Please see the main "mlr --help" for more information on syntax for these arguments.
-i {one of csv,dkvp,nidx,pprint,xtab}
--irs {record-separator character}
--ifs {field-separator character}
--ips {pair-separator character}
--repifs
--repips
--use-mmap
--no-mmap
Please see http://johnkerl.org/miller/doc/reference.html for more information including examples.
$ mlr --icsvlite --opprint cat data/join-left-example.csv id name 100 alice 200 bob 300 carol 400 david 500 edgar |
$ mlr --icsvlite --opprint cat data/join-right-example.csv status idcode present 400 present 100 missing 200 present 100 present 200 missing 100 missing 200 present 300 missing 600 present 400 present 400 present 300 present 100 missing 400 present 200 present 200 present 200 present 200 present 400 present 300 |
$ mlr --icsvlite --opprint join -u -j id -r idcode -f data/join-left-example.csv data/join-right-example.csv id name status 400 david present 100 alice present 200 bob missing 100 alice present 200 bob present 100 alice missing 200 bob missing 300 carol present 400 david present 400 david present 300 carol present 100 alice present 400 david missing 200 bob present 200 bob present 200 bob present 200 bob present 400 david present 300 carol present |
$ mlr --icsvlite --opprint sort -f idcode then join -j id -r idcode -f data/join-left-example.csv data/join-right-example.csv id name status 100 alice present 100 alice present 100 alice missing 100 alice present 200 bob missing 200 bob present 200 bob missing 200 bob present 200 bob present 200 bob present 200 bob present 300 carol present 300 carol present 300 carol present 400 david present 400 david present 400 david present 400 david missing 400 david present |
$ mlr --icsvlite --opprint join --np --ul --ur -u -j id -r idcode -f data/join-left-example.csv data/join-right-example.csv status idcode missing 600 id name 500 edgar |
$ mlr --csvlite --opprint cat data/self-join.csv data/self-join.csv a b c 1 2 3 1 4 5 1 2 3 1 4 5 |
$ mlr --csvlite --opprint join -j a --lp left_ --rp right_ -f data/self-join.csv data/self-join.csv a left_b left_c right_b right_c 1 2 3 2 3 1 4 5 2 3 1 2 3 4 5 1 4 5 4 5 |
$ mlr --csvlite --opprint join -j "" --lp left_ --rp right_ -f data/self-join.csv data/self-join.csv left_a left_b left_c right_a right_b right_c 1 2 3 1 2 3 1 4 5 1 2 3 1 2 3 1 4 5 1 4 5 1 4 5 |
label
$ mlr label --help
Usage: mlr label {new1,new2,new3,...}
Given n comma-separated names, renames the first n fields of each record to
have the respective name. (Fields past the nth are left with their original
names.) Particularly useful with --inidx, to give useful names to otherwise
integer-indexed fields.
% grep -v '^#' /etc/passwd | mlr --nidx --fs : --opprint label name,password,uid,gid,gecos,home_dir,shell | head name password uid gid gecos home_dir shell nobody * -2 -2 Unprivileged User /var/empty /usr/bin/false root * 0 0 System Administrator /var/root /bin/sh daemon * 1 1 System Services /var/root /usr/bin/false _uucp * 4 4 Unix to Unix Copy Protocol /var/spool/uucp /usr/sbin/uucico _taskgated * 13 13 Task Gate Daemon /var/empty /usr/bin/false _networkd * 24 24 Network Services /var/networkd /usr/bin/false _installassistant * 25 25 Install Assistant /var/empty /usr/bin/false _lp * 26 26 Printing Services /var/spool/cups /usr/bin/false _postfix * 27 27 Postfix Mail Server /var/spool/postfix /usr/bin/false
put
$ mlr put --help
Usage: mlr put [-v] {expression}
Adds/updates specified field(s).
With -v, first prints the AST (abstract syntax tree) for the expression, which
gives full transparency on the precedence and associativity rules of Miller's grammar.
Please use a dollar sign for field names and double-quotes for string literals.
Miller built-in variables are NF NR FNR FILENUM FILENAME PI E.
Multiple assignments may be separated with a semicolon.
Examples:
mlr put '$y = log10($x); $z = sqrt($y)'
mlr put '$filename = FILENAME'
mlr put '$colored_shape = $color . "_" . $shape'
mlr put '$y = cos($theta); $z = atan2($y, $x)'
Please see http://johnkerl.org/miller/doc/reference.html for more information including function list.
$ ruby -e '10.times{|i|puts "i=#{i}"}' | mlr --opprint put '$j=$i+1;$k=$i+$j'
i j k
0 1.000000 1.000000
1 2.000000 3.000000
2 3.000000 5.000000
3 4.000000 7.000000
4 5.000000 9.000000
5 6.000000 11.000000
6 7.000000 13.000000
7 8.000000 15.000000
8 9.000000 17.000000
9 10.000000 19.000000
$ mlr --opprint put '$nf=NF; $nr=NR; $fnr=FNR; $filenum=FILENUM; $filename=FILENAME' data/small data/small2 a b i x y nf nr fnr filenum filename pan pan 1 0.3467901443380824 0.7268028627434533 5 1 1 1 data/small eks pan 2 0.7586799647899636 0.5221511083334797 5 2 2 1 data/small wye wye 3 0.20460330576630303 0.33831852551664776 5 3 3 1 data/small eks wye 4 0.38139939387114097 0.13418874328430463 5 4 4 1 data/small wye pan 5 0.5732889198020006 0.8636244699032729 5 5 5 1 data/small pan eks 9999 0.267481232652199086 0.557077185510228001 5 6 1 2 data/small2 wye eks 10000 0.734806020620654365 0.884788571337605134 5 7 2 2 data/small2 pan wye 10001 0.870530722602517626 0.009854780514656930 5 8 3 2 data/small2 hat wye 10002 0.321507044286237609 0.568893318795083758 5 9 4 2 data/small2 pan zee 10003 0.272054845593895200 0.425789896597056627 5 10 5 2 data/small2
mlr --opprint put ' $nf = NF; $nr = NR; $fnr = FNR; $filenum = FILENUM; $filename = FILENAME' \ data/small data/small2
regularize
$ mlr regularize --help Usage: mlr regularize For records seen earlier in the data stream with same field names in a different order, outputs them with field names in the previously encountered order. Example: input records a=1,c=2,b=3, then e=4,d=5, then c=7,a=6,b=8 output as a=1,c=2,b=3, then e=4,d=5, then a=6,c=7,b=8
rename
$ mlr rename --help
Usage: mlr rename {old1,new1,old2,new2,...}
Renames specified fields.
$ mlr --opprint cat data/small a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 wye pan 5 0.5732889198020006 0.8636244699032729 |
$ mlr --opprint rename i,INDEX,b,COLUMN2 data/small a COLUMN2 INDEX x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 wye pan 5 0.5732889198020006 0.8636244699032729 |
$ sed 's/y/COLUMN5/g' data/small a=pan,b=pan,i=1,x=0.3467901443380824,COLUMN5=0.7268028627434533 a=eks,b=pan,i=2,x=0.7586799647899636,COLUMN5=0.5221511083334797 a=wCOLUMN5e,b=wCOLUMN5e,i=3,x=0.20460330576630303,COLUMN5=0.33831852551664776 a=eks,b=wCOLUMN5e,i=4,x=0.38139939387114097,COLUMN5=0.13418874328430463 a=wCOLUMN5e,b=pan,i=5,x=0.5732889198020006,COLUMN5=0.8636244699032729 |
$ mlr rename y,COLUMN5 data/small a=pan,b=pan,i=1,x=0.3467901443380824,COLUMN5=0.7268028627434533 a=eks,b=pan,i=2,x=0.7586799647899636,COLUMN5=0.5221511083334797 a=wye,b=wye,i=3,x=0.20460330576630303,COLUMN5=0.33831852551664776 a=eks,b=wye,i=4,x=0.38139939387114097,COLUMN5=0.13418874328430463 a=wye,b=pan,i=5,x=0.5732889198020006,COLUMN5=0.8636244699032729 |
reorder
$ mlr reorder --help
Usage: mlr reorder [options]
-f {a,b,c} Field names to reorder.
-e Put specified field names at record end: default is to put at record start.
Example: mlr reorder -f a,b sends input record "d=4,b=2,a=1,c=3" to "a=1,b=2,d=4,c=3".
Example: mlr reorder -e -f a,b sends input record "d=4,b=2,a=1,c=3" to "d=4,c=3,a=1,b=2".
$ mlr --opprint cat data/small a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 wye pan 5 0.5732889198020006 0.8636244699032729 | |
$ mlr --opprint reorder -f i,b data/small i b a x y 1 pan pan 0.3467901443380824 0.7268028627434533 2 pan eks 0.7586799647899636 0.5221511083334797 3 wye wye 0.20460330576630303 0.33831852551664776 4 wye eks 0.38139939387114097 0.13418874328430463 5 pan wye 0.5732889198020006 0.8636244699032729 |
$ mlr --opprint reorder -e -f i,b data/small a x y i b pan 0.3467901443380824 0.7268028627434533 1 pan eks 0.7586799647899636 0.5221511083334797 2 pan wye 0.20460330576630303 0.33831852551664776 3 wye eks 0.38139939387114097 0.13418874328430463 4 wye wye 0.5732889198020006 0.8636244699032729 5 pan |
sort
$ mlr sort --help
Usage: mlr sort {flags}
Flags:
-f {comma-separated field names} Lexical ascending
-n {comma-separated field names} Numerical ascending; nulls sort last
-nf {comma-separated field names} Numerical ascending; nulls sort last
-r {comma-separated field names} Lexical descending
-nr {comma-separated field names} Numerical descending; nulls sort first
Sorts records primarily by the first specified field, secondarily by the second field, and so on.
Example:
mlr sort -f a,b -nr x,y,z
which is the same as:
mlr sort -f a -f b -nr x -nr y -nr z
$ mlr --opprint sort -f a -nr x data/small a b i x y eks pan 2 0.7586799647899636 0.5221511083334797 eks wye 4 0.38139939387114097 0.13418874328430463 pan pan 1 0.3467901443380824 0.7268028627434533 wye pan 5 0.5732889198020006 0.8636244699032729 wye wye 3 0.20460330576630303 0.33831852551664776
$ head -n 10 data/multicountdown.dat upsec=0.002,color=green,count=1203 upsec=0.083,color=red,count=3817 upsec=0.188,color=red,count=3801 upsec=0.395,color=blue,count=2697 upsec=0.526,color=purple,count=953 upsec=0.671,color=blue,count=2684 upsec=0.899,color=purple,count=926 upsec=0.912,color=red,count=3798 upsec=1.093,color=blue,count=2662 upsec=1.327,color=purple,count=917
$ head -n 20 data/multicountdown.dat | mlr --opprint sort -f color upsec color count 0.395 blue 2697 0.671 blue 2684 1.093 blue 2662 2.064 blue 2659 2.2880000000000003 blue 2647 0.002 green 1203 1.407 green 1187 1.448 green 1177 2.313 green 1161 0.526 purple 953 0.899 purple 926 1.327 purple 917 1.703 purple 908 0.083 red 3817 0.188 red 3801 0.912 red 3798 1.416 red 3788 1.587 red 3782 1.601 red 3755 1.832 red 3717
stats1
$ mlr stats1 --help
Usage: mlr stats1 [options]
Options:
-a {sum,count,...} Names of accumulators: p10 p25.2 p50 p98 p100 etc. and/or one or more of
count mode sum mean stddev var meaneb min max
-f {a,b,c} Value-field names on which to compute statistics
-g {d,e,f} Optional group-by-field names
Example: mlr stats1 -a min,p10,p50,p90,max -f value -g size,shape
Example: mlr stats1 -a count,mode -f size
Example: mlr stats1 -a count,mode -f size -g shape
Notes:
* p50 is a synonym for median.
* min and max output the same results as p0 and p100, respectively, but use less memory.
* count and mode allow text input; the rest require numeric input. In particular, 1 and 1.0
are distinct text for count and mode.
* When there are mode ties, the first-encountered datum wins.
$ mlr --oxtab stats1 -a count,sum,min,p10,p50,mean,p90,max -f x,y data/medium x_count 10000 x_sum 4986.019682 x_min 0.000045 x_p10 0.093322 x_p50 0.501159 x_mean 0.498602 x_p90 0.900794 x_max 0.999953 y_count 10000 y_sum 5062.057445 y_min 0.000088 y_p10 0.102132 y_p50 0.506021 y_mean 0.506206 y_p90 0.905366 y_max 0.999965 |
$ mlr --opprint stats1 -a mean -f x,y -g b then sort -f b data/medium b x_mean y_mean eks 0.506361 0.510293 hat 0.487899 0.513118 pan 0.497304 0.499599 wye 0.497593 0.504596 zee 0.504242 0.502997 |
$ mlr --opprint stats1 -a p50,p99 -f u,v -g color then put '$ur=$u_p99/$u_p50;$vr=$v_p99/$v_p50' data/colored-shapes.dkvp color u_p50 u_p99 v_p50 v_p99 ur vr green 0.505848 0.990764 0.500787 0.989642 1.958620 1.976174 red 0.500564 0.989587 0.500415 0.996280 1.976944 1.990908 yellow 0.506272 0.989401 0.502912 0.989215 1.954287 1.966974 blue 0.506270 0.990783 0.490120 0.990957 1.957025 2.021866 purple 0.500051 0.990108 0.505464 0.990086 1.980014 1.958767 orange 0.488821 0.991091 0.491891 0.991451 2.027513 2.015591 |
$ mlr --opprint count-distinct -f shape then sort -nr count data/colored-shapes.dkvp shape count square 41588 triangle 33477 circle 24935 |
$ mlr --opprint stats1 -a mode -f color -g shape data/colored-shapes.dkvp shape color_mode circle red square red triangle red |
stats2
$ mlr stats2 --help
Usage: mlr stats2 [options]
-a {linreg-ols,corr,...} Names of accumulators: one or more of
linreg-pca linreg-ols r2 corr cov covx
r2 is a quality metric for linreg-ols; linrec-pca outputs its own quality metric.
-f {a,b,c,d} Value-field name-pairs on which to compute statistics.
There must be an even number of names.
-g {e,f,g} Optional group-by-field names.
-v Print additional output for linreg-pca.
Example: mlr stats2 -a linreg-pca -f x,y
Example: mlr stats2 -a linreg-ols,r2 -f x,y -g size,shape
Example: mlr stats2 -a corr -f x,y
$ mlr --oxtab put '$x2=$x*$x; $xy=$x*$y; $y2=$y**2' then stats2 -a cov,corr -f x,y,y,y,x2,xy,x2,y2 data/medium x_y_cov 0.000043 x_y_corr 0.000504 y_y_cov 0.084611 y_y_corr 1.000000 x2_xy_cov 0.041884 x2_xy_corr 0.630174 x2_y2_cov -0.000310 x2_y2_corr -0.003425 |
$ mlr --opprint put '$x2=$x*$x; $xy=$x*$y; $y2=$y**2' then stats2 -a linreg-ols,r2 -f x,y,y,y,xy,y2 -g a data/medium a x_y_ols_m x_y_ols_b x_y_ols_n x_y_r2 y_y_ols_m y_y_ols_b y_y_ols_n y_y_r2 xy_y2_ols_m xy_y2_ols_b xy_y2_ols_n xy_y2_r2 pan 0.017026 0.500403 2081 0.000287 1.000000 0.000000 2081 1.000000 0.878132 0.119082 2081 0.417498 eks 0.040780 0.481402 1965 0.001646 1.000000 0.000000 1965 1.000000 0.897873 0.107341 1965 0.455632 wye -0.039153 0.525510 1966 0.001505 1.000000 0.000000 1966 1.000000 0.853832 0.126745 1966 0.389917 zee 0.002781 0.504307 2047 0.000008 1.000000 0.000000 2047 1.000000 0.852444 0.124017 2047 0.393566 hat -0.018621 0.517901 1941 0.000352 1.000000 0.000000 1941 1.000000 0.841230 0.135573 1941 0.368794 |
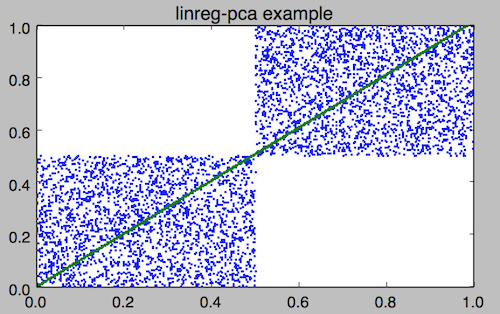
mlr filter '($x<.5 && $y<.5) || ($x>.5 && $y>.5)' data/medium > data/medium-squares mlr --ofs newline stats2 -a linreg-pca -f x,y data/medium-squares x_y_pca_m=1.014419 x_y_pca_b=0.000308 x_y_pca_quality=0.861354 # Set x_y_pca_m and x_y_pca_b as shell variables eval $(mlr --ofs newline stats2 -a linreg-pca -f x,y data/medium-squares) # In addition to x and y, make a new yfit which is the line fit. Plot using your favorite tool. mlr --onidx put '$yfit='$x_y_pca_m'*$x+'$x_y_pca_b then cut -x -f a,b,i data/medium-squares \ | pgr -p -title 'linreg-pca example' -xmin 0 -xmax 1 -ymin 0 -ymax 1
$ head -n 10 data/multicountdown.dat upsec=0.002,color=green,count=1203 upsec=0.083,color=red,count=3817 upsec=0.188,color=red,count=3801 upsec=0.395,color=blue,count=2697 upsec=0.526,color=purple,count=953 upsec=0.671,color=blue,count=2684 upsec=0.899,color=purple,count=926 upsec=0.912,color=red,count=3798 upsec=1.093,color=blue,count=2662 upsec=1.327,color=purple,count=917
$ mlr --oxtab stats2 -a linreg-pca -f upsec,count -g color then put '$donesec = -$upsec_count_pca_b/$upsec_count_pca_m' data/multicountdown.dat color green upsec_count_pca_m -32.756917 upsec_count_pca_b 1213.722730 upsec_count_pca_n 24 upsec_count_pca_quality 0.999984 donesec 37.052410 color red upsec_count_pca_m -37.367646 upsec_count_pca_b 3810.133400 upsec_count_pca_n 30 upsec_count_pca_quality 0.999989 donesec 101.963431 color blue upsec_count_pca_m -29.231212 upsec_count_pca_b 2698.932820 upsec_count_pca_n 25 upsec_count_pca_quality 0.999959 donesec 92.330514 color purple upsec_count_pca_m -39.030097 upsec_count_pca_b 979.988341 upsec_count_pca_n 21 upsec_count_pca_quality 0.999991 donesec 25.108529
step
$ mlr step --help
Usage: mlr step [options]
-a {delta,rsum,...} Names of steppers: one or more of
delta rsum counter
-f {a,b,c} Value-field names on which to compute statistics
-g {d,e,f} Group-by-field names
Computes values dependent on the previous record, optionally grouped by category.
$ mlr --opprint step -a delta,rsum,counter -f x data/medium | head -15 a b i x y x_delta x_rsum x_counter pan pan 1 0.3467901443380824 0.7268028627434533 0.346790 0.346790 1 eks pan 2 0.7586799647899636 0.5221511083334797 0.411890 1.105470 2 wye wye 3 0.20460330576630303 0.33831852551664776 -0.554077 1.310073 3 eks wye 4 0.38139939387114097 0.13418874328430463 0.176796 1.691473 4 wye pan 5 0.5732889198020006 0.8636244699032729 0.191890 2.264762 5 zee pan 6 0.5271261600918548 0.49322128674835697 -0.046163 2.791888 6 eks zee 7 0.6117840605678454 0.1878849191181694 0.084658 3.403672 7 zee wye 8 0.5985540091064224 0.976181385699006 -0.013230 4.002226 8 hat wye 9 0.03144187646093577 0.7495507603507059 -0.567112 4.033668 9 pan wye 10 0.5026260055412137 0.9526183602969864 0.471184 4.536294 10 pan pan 11 0.7930488423451967 0.6505816637259333 0.290423 5.329343 11 zee pan 12 0.3676141320555616 0.23614420670296965 -0.425435 5.696957 12 eks pan 13 0.4915175580479536 0.7709126592971468 0.123903 6.188474 13 eks zee 14 0.5207382318405251 0.34141681118811673 0.029221 6.709213 14 |
$ mlr --opprint step -a delta,rsum,counter -f x -g a data/medium | head -15 a b i x y x_delta x_rsum x_counter pan pan 1 0.3467901443380824 0.7268028627434533 0.346790 0.346790 1 eks pan 2 0.7586799647899636 0.5221511083334797 0.758680 0.758680 1 wye wye 3 0.20460330576630303 0.33831852551664776 0.204603 0.204603 1 eks wye 4 0.38139939387114097 0.13418874328430463 -0.377281 1.140079 2 wye pan 5 0.5732889198020006 0.8636244699032729 0.368686 0.777892 2 zee pan 6 0.5271261600918548 0.49322128674835697 0.527126 0.527126 1 eks zee 7 0.6117840605678454 0.1878849191181694 0.230385 1.751863 3 zee wye 8 0.5985540091064224 0.976181385699006 0.071428 1.125680 2 hat wye 9 0.03144187646093577 0.7495507603507059 0.031442 0.031442 1 pan wye 10 0.5026260055412137 0.9526183602969864 0.155836 0.849416 2 pan pan 11 0.7930488423451967 0.6505816637259333 0.290423 1.642465 3 zee pan 12 0.3676141320555616 0.23614420670296965 -0.230940 1.493294 3 eks pan 13 0.4915175580479536 0.7709126592971468 -0.120267 2.243381 4 eks zee 14 0.5207382318405251 0.34141681118811673 0.029221 2.764119 5 |
$ each 10 uptime | mlr -p step -a delta -f 11 ... 20:08 up 36 days, 10:38, 5 users, load averages: 1.42 1.62 1.73 0.000000 20:08 up 36 days, 10:38, 5 users, load averages: 1.55 1.64 1.74 0.020000 20:08 up 36 days, 10:38, 7 users, load averages: 1.58 1.65 1.74 0.010000 20:08 up 36 days, 10:38, 9 users, load averages: 1.78 1.69 1.76 0.040000 20:08 up 36 days, 10:39, 9 users, load averages: 2.12 1.76 1.78 0.070000 20:08 up 36 days, 10:39, 9 users, load averages: 2.51 1.85 1.81 0.090000 20:08 up 36 days, 10:39, 8 users, load averages: 2.79 1.92 1.83 0.070000 20:08 up 36 days, 10:39, 4 users, load averages: 2.64 1.90 1.83 -0.020000
tac
$ mlr tac --help Usage: mlr tac Prints records in reverse order from the order in which they were encountered.
$ mlr --icsv --opprint cat a.csv a b c 1 2 3 4 5 6 |
$ mlr --icsv --opprint cat b.csv a b c 7 8 9 |
$ mlr --icsv --opprint tac a.csv b.csv a b c 7 8 9 4 5 6 1 2 3 |
$ mlr --icsv --opprint put '$filename=FILENAME' then tac a.csv b.csv a b c filename 7 8 9 b.csv 4 5 6 a.csv 1 2 3 a.csv |
tail
$ mlr tail --help
Usage: mlr tail [options]
-n {count} Tail count to print; default 10
-g {a,b,c} Optional group-by-field names for tail counts
Passes through the last n records, optionally by category.
$ mlr --opprint tail -n 4 data/colored-shapes.dkvp color shape flag i u v w x yellow circle 1 99997 0.5228034832314841 0.7478634261534541 0.49477944033468396 6.085638633037881 red triangle 0 99998 0.8566019561040149 0.5583785393850178 0.4993735796215503 6.393409471109115 yellow triangle 1 99999 0.5369350176939407 0.5197619334387739 0.5064468446479313 3.2682256831629695 green square 0 100000 0.0277485352321325 0.5303062901341336 0.5274344049261097 5.806843329974349 |
$ mlr --opprint tail -n 1 -g shape data/colored-shapes.dkvp color shape flag i u v w x yellow circle 1 99997 0.5228034832314841 0.7478634261534541 0.49477944033468396 6.085638633037881 green square 0 100000 0.0277485352321325 0.5303062901341336 0.5274344049261097 5.806843329974349 yellow triangle 1 99999 0.5369350176939407 0.5197619334387739 0.5064468446479313 3.2682256831629695 |
top
$ mlr top --help
Usage: mlr top [options]
-f {a,b,c} Value-field names for top counts
-g {d,e,f} Optional group-by-field names for top counts
-n {count} How many records to print per category; default 1
-a Print all fields for top-value records; default is
to print only value and group-by fields.
--min Print top smallest values; default is top largest values
Prints the n records with smallest/largest values at specified fields, optionally by category.
$ mlr --opprint top -n 4 -f x data/medium top_idx x_top 1 0.999953 2 0.999823 3 0.999733 4 0.999563 |
$ mlr --opprint top -n 2 -f x -g a then sort -f a data/medium a top_idx x_top eks 1 0.998811 eks 2 0.998534 hat 1 0.999953 hat 2 0.999733 pan 1 0.999403 pan 2 0.999044 wye 1 0.999823 wye 2 0.999264 zee 1 0.999490 zee 2 0.999438 |
uniq
$ mlr uniq --help
Usage: mlr uniq [options]
-g {d,e,f} Group-by-field names for uniq counts
-c Show repeat counts in addition to unique values
Prints distinct values for specified field names. With -c, same as count-distinct.
$ wc -l data/colored-shapes.dkvp 100000 data/colored-shapes.dkvp |
$ mlr uniq -g color,shape data/colored-shapes.dkvp color=green,shape=circle color=red,shape=square color=yellow,shape=circle color=red,shape=circle color=blue,shape=circle color=purple,shape=triangle color=blue,shape=triangle color=green,shape=square color=red,shape=triangle color=yellow,shape=triangle color=purple,shape=square color=blue,shape=square color=yellow,shape=square color=green,shape=triangle color=purple,shape=circle color=orange,shape=triangle color=orange,shape=square color=orange,shape=circle |
$ mlr --opprint uniq -g color,shape -c then sort -f color,shape data/colored-shapes.dkvp color shape count blue circle 3578 blue square 6016 blue triangle 4843 green circle 2832 green square 4678 green triangle 3924 orange circle 705 orange square 1196 orange triangle 954 purple circle 2861 purple square 4808 purple triangle 3841 red circle 11477 red square 19051 red triangle 15248 yellow circle 3482 yellow square 5839 yellow triangle 4667 |
Functions for filter and put
$ mlr --help-all-functions abs (math: #args=1): Absolute value. acos (math: #args=1): Inverse trigonometric cosine. acosh (math: #args=1): Inverse hyperbolic cosine. asin (math: #args=1): Inverse trigonometric sine. asinh (math: #args=1): Inverse hyperbolic sine. atan (math: #args=1): One-argument arctangent. atan2 (math: #args=2): Two-argument arctangent. atanh (math: #args=1): Inverse hyperbolic tangent. cbrt (math: #args=1): Cube root. ceil (math: #args=1): Ceiling: nearest integer at or above. cos (math: #args=1): Trigonometric cosine. cosh (math: #args=1): Hyperbolic cosine. erf (math: #args=1): Error function. erfc (math: #args=1): Complementary error function. exp (math: #args=1): Exponential function e**x. expm1 (math: #args=1): e**x - 1. floor (math: #args=1): Floor: nearest integer at or below. invqnorm (math: #args=1): Inverse of normal cumulative distribution function. Note that invqorm(urand()) is normally distributed. log (math: #args=1): Natural (base-e) logarithm. log10 (math: #args=1): Base-10 logarithm. log1p (math: #args=1): log(1-x). max (math: #args=2): max of two numbers; null loses min (math: #args=2): min of two numbers; null loses pow (math: #args=2): Exponentiation; same as **. qnorm (math: #args=1): Normal cumulative distribution function. round (math: #args=1): Round to nearest integer. roundm (math: #args=2): Round to nearest multiple of m: roundm($x,$m) is the same as round($x/$m)*$m sin (math: #args=1): Trigonometric sine. sinh (math: #args=1): Hyperbolic sine. sqrt (math: #args=1): Square root. tan (math: #args=1): Trigonometric tangent. tanh (math: #args=1): Hyperbolic tangent. urand (math: #args=0): Floating-point numbers on the unit interval. Int-valued example: '$n=floor(20+urand()*11)'. + (math: #args=2): Addition. - (math: #args=1): Unary minus. - (math: #args=2): Subtraction. * (math: #args=2): Multiplication. / (math: #args=2): Division. % (math: #args=2): Remainder; never negative-valued. ** (math: #args=2): Exponentiation; same as pow. == (boolean: #args=2): String/numeric equality. Mixing number and string results in string compare. != (boolean: #args=2): String/numeric inequality. Mixing number and string results in string compare. > (boolean: #args=2): String/numeric greater-than. Mixing number and string results in string compare. >= (boolean: #args=2): String/numeric greater-than-or-equals. Mixing number and string results in string compare. < (boolean: #args=2): String/numeric less-than. Mixing number and string results in string compare. <= (boolean: #args=2): String/numeric less-than-or-equals. Mixing number and string results in string compare. && (boolean: #args=2): Logical AND. || (boolean: #args=2): Logical OR. ! (boolean: #args=1): Logical negation. strlen (string: #args=1): String length. sub (string: #args=3): Example: '$name=sub($name, "old", "new")'. Regexes not supported. tolower (string: #args=1): Convert string to lowercase. toupper (string: #args=1): Convert string to uppercase. . (string: #args=2): String concatenation. boolean (conversion: #args=1): Convert int/float/bool/string to boolean. float (conversion: #args=1): Convert int/float/bool/string to float. int (conversion: #args=1): Convert int/float/bool/string to int. string (conversion: #args=1): Convert int/float/bool/string to string. hexfmt (conversion: #args=1): Convert int to string, e.g. 255 to "0xff". fmtnum (conversion: #args=2): Convert int/float/bool to string using printf-style format string, e.g. "%06lld". gmt2sec (time: #args=1): Parses GMT timestamp as integer seconds since epoch. sec2gmt (time: #args=1): Formats seconds since epoch (integer part only) as GMT timestamp. systime (time: #args=0): Floating-point seconds since the epoch. To set the seed for urand, you may specify decimal or hexadecimal 32-bit numbers of the form "mlr --seed 123456789" or "mlr --seed 0xcafefeed". Miller's built-in variables are NF, NR, FNR, FILENUM, and FILENAME (awk-like) along with the mathematical constants PI and E.
Data types
Miller’s input and output are all string-oriented: there is (as of August 2015 anyway) no support for binary record packing. In this sense, everything is a string in and out of Miller. During processing, field names are always strings, even if they have names like "3"; field values are usually strings. Field values’ ability to be interpreted as a non-string type only has meaning when comparison or function operations are done on them. And it is an error condition if Miller encounters non-numeric (or otherwise mistyped) data in a field in which it has been asked to do numeric (or otherwise type-specific) operations. Field values are treated as numeric for the following:-
Numeric sort: mlr sort -n, mlr sort -nr.
Statistics: mlr histogram, mlr stats1, mlr stats2.
Cross-record arithmetic: mlr step.
-
Miller’s types for function processing are null (empty string), error, string, float (double-precision), int (64-bit signed), and boolean.
On input, string values representable as numbers (e.g. "3" or "3.1") are treated as float. If a record
has x=1,y=2 then mlr put '$z=$x+$y' will produce x=1,y=2,z=3, and
mlr put '$z=$x.$y' gives an error. To coerce back to string for
processing, use the string function:
mlr put '$z=string($x).string($y)' will produce x=1,y=2,z=12.
On input, string values representable as boolean (e.g. "true",
"false") are not automatically treated as boolean.
(This is because "true" and "false" are ordinary words, and auto string-to-boolean
on a column consisting of words would result in some strings mixed with some booleans.)
Use the boolean function to coerce: e.g. giving the record x=1,y=2,w=false to
mlr put '$z=($x<$y) || boolean($w)'.
Functions take types as described in mlr --help-all-functions: for example, log10
takes float input and produces float output, gmt2sec maps string to int, and sec2gmt
maps int to string.
All math functions described in mlr --help-all-functions take integer as well as float input.
Null data
One of Miller’s key features is its support for heterogeneous data. Accordingly, if you try to sort on field hostname when not all records in the data stream have a field named hostname, it is not an error (although you could pre-filter the data stream using mlr having-fields --at-least hostname then sort ...). Rather, records lacking one or more sort keys are simply output contiguously by mlr sort. Field values may also be null by being specified with present key but empty value: e.g. sending x=,y=2 to mlr put '$z=$x+$y'. Rules for null-handling:- Records with one or more null sort-field values sort after records with all sort-field values present:
$ mlr --opprint cat data/sort-null.dat a b 3 2 1 8 - 4 5 7
$ mlr --opprint sort -n a data/sort-null.dat a b 1 8 3 2 5 7 - 4
$ mlr --opprint sort -nr a data/sort-null.dat a b - 4 5 7 3 2 1 8
- Functions which have one or more null arguments produce null output: e.g.
$ echo 'x=2,y=3' | mlr put '$a=$x+$y' x=2,y=3,a=5.000000
$ echo 'x=,y=3' | mlr put '$a=$x+$y' x=,y=3,a=
$ echo 'x=,y=3' | mlr put '$a=log($x);$b=log($y)' x=,y=3,a=,b=1.098612
- The min and max functions are special: if one argument is non-null, it wins:
$ echo 'x=,y=3' | mlr put '$a=min($x,$y);$b=max($x,$y)' x=,y=3,a=3.000000,b=3.000000