Overview:
• About Miller
• Miller in 10 minutes
• File formats
• Miller features in the context of the Unix toolkit
• Record-heterogeneity
• Internationalization
Using Miller:
• FAQ
• Sharing data with other languages
• Cookbook part 1
• Cookbook part 2
• Cookbook part 3
• Data-diving examples
• Manpage
• Reference
• Reference: Verbs
• Reference: DSL
• Documents by release
• Installation, portability, dependencies, and testing
Background:
• Why?
• Why C?
• Why call it Miller?
• How original is Miller?
• Performance
Repository:
• Things to do
• Contact information
• GitHub repo
• altkv
• bar
• bootstrap
• cat
• check
• clean-whitespace
• count-distinct
• count-similar
• cut
• decimate
• fill-down
• filter
• Features which filter shares with put
• fraction
• grep
• group-by
• group-like
• having-fields
• head
• histogram
• join
• label
• least-frequent
• merge-fields
• most-frequent
• nest
• nothing
• put
• Features which put shares with filter
• regularize
• rename
• reorder
• repeat
• reshape
• sample
• sec2gmt
• sec2gmtdate
• seqgen
• shuffle
• sort
• stats1
• stats2
• step
• tac
• tail
• tee
• top
• uniq
• unsparsify
Overview
When you type mlr {something} myfile.dat, the {something}
part is called a verb. It specifies how you want to transform your data.
(See also here for a breakdown.)
The following is an alphabetical list of verbs with their descriptions.
The verbs put and filter are special in that they have a
rich expression language (domain-specific language, or “DSL”).
More information about them can be found here.
Here’s a comparison of verbs and put/filter DSL expressions:
Example:
$ mlr stats1 -a sum -f x -g a data/small a=pan,x_sum=0.346790 a=eks,x_sum=1.140079 a=wye,x_sum=0.777892
|
Example:
$ mlr put -q '@x_sum[$a] += $x; end{emit @x_sum, "a"}' data/small
a=pan,x_sum=0.346790
a=eks,x_sum=1.140079
a=wye,x_sum=0.777892
|
altkv
Map list of values to alternating key/value pairs.
$ mlr altkv -h Usage: mlr altkv [no options] Given fields with values of the form a,b,c,d,e,f emits a=b,c=d,e=f pairs.
$ echo 'a,b,c,d,e,f' | mlr altkv a=b,c=d,e=f
$ echo 'a,b,c,d,e,f,g' | mlr altkv a=b,c=d,e=f,4=g
bar
Cheesy bar-charting.
$ mlr bar -h
Usage: mlr bar [options]
Replaces a numeric field with a number of asterisks, allowing for cheesy
bar plots. These align best with --opprint or --oxtab output format.
Options:
-f {a,b,c} Field names to convert to bars.
-c {character} Fill character: default '*'.
-x {character} Out-of-bounds character: default '#'.
-b {character} Blank character: default '.'.
--lo {lo} Lower-limit value for min-width bar: default '0.000000'.
--hi {hi} Upper-limit value for max-width bar: default '100.000000'.
-w {n} Bar-field width: default '40'.
--auto Automatically computes limits, ignoring --lo and --hi.
Holds all records in memory before producing any output.
$ mlr --opprint cat data/small a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 wye pan 5 0.5732889198020006 0.8636244699032729
$ mlr --opprint bar --lo 0 --hi 1 -f x,y data/small a b i x y pan pan 1 *************........................... *****************************........... eks pan 2 ******************************.......... ********************.................... wye wye 3 ********................................ *************........................... eks wye 4 ***************......................... *****................................... wye pan 5 **********************.................. **********************************......
$ mlr --opprint bar --lo 0.4 --hi 0.6 -f x,y data/small a b i x y pan pan 1 #....................................... ***************************************# eks pan 2 ***************************************# ************************................ wye wye 3 #....................................... #....................................... eks wye 4 #....................................... #....................................... wye pan 5 **********************************...... ***************************************#
$ mlr --opprint bar --auto -f x,y data/small a b i x y pan pan 1 [0.204603]**********..............................[0.75868] [0.134189]********************************........[0.863624] eks pan 2 [0.204603]***************************************#[0.75868] [0.134189]*********************...................[0.863624] wye wye 3 [0.204603]#.......................................[0.75868] [0.134189]***********.............................[0.863624] eks wye 4 [0.204603]************............................[0.75868] [0.134189]#.......................................[0.863624] wye pan 5 [0.204603]**************************..............[0.75868] [0.134189]***************************************#[0.863624]
bootstrap
$ mlr bootstrap --help
Usage: mlr bootstrap [options]
Emits an n-sample, with replacement, of the input records.
Options:
-n {number} Number of samples to output. Defaults to number of input records.
Must be non-negative.
See also mlr sample and mlr shuffle.
$ mlr --opprint stats1 -a mean,count -f u -g color data/colored-shapes.dkvp color u_mean u_count yellow 0.497129 1413 red 0.492560 4641 purple 0.494005 1142 green 0.504861 1109 blue 0.517717 1470 orange 0.490532 303 $ mlr --opprint bootstrap then stats1 -a mean,count -f u -g color data/colored-shapes.dkvp color u_mean u_count yellow 0.500651 1380 purple 0.501556 1111 green 0.503272 1068 red 0.493895 4702 blue 0.512529 1496 orange 0.521030 321 $ mlr --opprint bootstrap then stats1 -a mean,count -f u -g color data/colored-shapes.dkvp color u_mean u_count yellow 0.498046 1485 blue 0.513576 1417 red 0.492870 4595 orange 0.507697 307 green 0.496803 1075 purple 0.486337 1199 $ mlr --opprint bootstrap then stats1 -a mean,count -f u -g color data/colored-shapes.dkvp color u_mean u_count blue 0.522921 1447 red 0.490717 4617 yellow 0.496450 1419 purple 0.496523 1192 green 0.507569 1111 orange 0.468014 292
cat
Most useful for format conversions (see
File formats), and concatenating multiple
same-schema CSV files to have the same header:
$ mlr cat -h
Usage: mlr cat [options]
Passes input records directly to output. Most useful for format conversion.
Options:
-n Prepend field "n" to each record with record-counter starting at 1
-g {comma-separated field name(s)} When used with -n/-N, writes record-counters
keyed by specified field name(s).
-N {name} Prepend field {name} to each record with record-counter starting at 1
$ cat data/a.csv a,b,c 1,2,3 4,5,6 |
$ cat data/b.csv a,b,c 7,8,9 |
$ mlr --csv cat data/a.csv data/b.csv a,b,c 1,2,3 4,5,6 7,8,9 |
$ mlr --icsv --oxtab cat data/a.csv data/b.csv a 1 b 2 c 3 a 4 b 5 c 6 a 7 b 8 c 9 |
$ mlr --csv cat -n data/a.csv data/b.csv n,a,b,c 1,1,2,3 2,4,5,6 3,7,8,9 |
$ mlr --opprint cat data/small a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 wye pan 5 0.5732889198020006 0.8636244699032729 |
$ mlr --opprint cat -n -g a data/small n a b i x y 1 pan pan 1 0.3467901443380824 0.7268028627434533 1 eks pan 2 0.7586799647899636 0.5221511083334797 1 wye wye 3 0.20460330576630303 0.33831852551664776 2 eks wye 4 0.38139939387114097 0.13418874328430463 2 wye pan 5 0.5732889198020006 0.8636244699032729 |
check
$ mlr check --help Usage: mlr check Consumes records without printing any output. Useful for doing a well-formatted check on input data.
clean-whitespace
$ mlr clean-whitespace --help
Usage: mlr clean-whitespace [options] {old1,new1,old2,new2,...}
For each record, for each field in the record, whitespace-cleans the keys and
values. Whitespace-cleaning entails stripping leading and trailing whitespace,
and replacing multiple whitespace with singles. For finer-grained control,
please see the DSL functions lstrip, rstrip, strip, collapse_whitespace,
and clean_whitespace.
Options:
-k|--keys-only Do not touch values.
-v|--values-only Do not touch keys.
It is an error to specify -k as well as -v.
$ mlr --icsv --ojson cat data/clean-whitespace.csv
{ " Name ": " Ann Simons", " Preference ": " blue " }
{ " Name ": "Bob Wang ", " Preference ": " red " }
{ " Name ": " Carol Vee", " Preference ": " yellow" }
$ mlr --icsv --ojson clean-whitespace -k data/clean-whitespace.csv
{ "Name": " Ann Simons", "Preference": " blue " }
{ "Name": "Bob Wang ", "Preference": " red " }
{ "Name": " Carol Vee", "Preference": " yellow" }
$ mlr --icsv --ojson clean-whitespace -v data/clean-whitespace.csv
{ " Name ": "Ann Simons", " Preference ": "blue" }
{ " Name ": "Bob Wang", " Preference ": "red" }
{ " Name ": "Carol Vee", " Preference ": "yellow" }
$ mlr --icsv --ojson clean-whitespace data/clean-whitespace.csv
{ "Name": "Ann Simons", "Preference": "blue" }
{ "Name": "Bob Wang", "Preference": "red" }
{ "Name": "Carol Vee", "Preference": "yellow" }
count-distinct
$ mlr count-distinct --help
Usage: mlr count-distinct [options]
Prints number of records having distinct values for specified field names.
Same as uniq -c.
Options:
-f {a,b,c} Field names for distinct count.
-n Show only the number of distinct values. Not compatible with -u.
-o {name} Field name for output count. Default "count".
Ignored with -u.
-u Do unlashed counts for multiple field names. With -f a,b and
without -u, computes counts for distinct combinations of a
and b field values. With -f a,b and with -u, computes counts
for distinct a field values and counts for distinct b field
values separately.
$ mlr count-distinct -f a,b then sort -nr count data/medium a=zee,b=wye,count=455 a=pan,b=eks,count=429 a=pan,b=pan,count=427 a=wye,b=hat,count=426 a=hat,b=wye,count=423 a=pan,b=hat,count=417 a=eks,b=hat,count=417 a=eks,b=eks,count=413 a=pan,b=zee,count=413 a=zee,b=hat,count=409 a=eks,b=wye,count=407 a=zee,b=zee,count=403 a=pan,b=wye,count=395 a=wye,b=pan,count=392 a=zee,b=eks,count=391 a=zee,b=pan,count=389 a=hat,b=eks,count=389 a=wye,b=eks,count=386 a=hat,b=zee,count=385 a=wye,b=zee,count=385 a=hat,b=hat,count=381 a=wye,b=wye,count=377 a=eks,b=pan,count=371 a=hat,b=pan,count=363 a=eks,b=zee,count=357
$ mlr count-distinct -u -f a,b data/medium field=a,value=pan,count=2081 field=a,value=eks,count=1965 field=a,value=wye,count=1966 field=a,value=zee,count=2047 field=a,value=hat,count=1941 field=b,value=pan,count=1942 field=b,value=wye,count=2057 field=b,value=zee,count=1943 field=b,value=eks,count=2008 field=b,value=hat,count=2050
$ mlr count-distinct -f a,b -o someothername then sort -nr someothername data/medium a=zee,b=wye,someothername=455 a=pan,b=eks,someothername=429 a=pan,b=pan,someothername=427 a=wye,b=hat,someothername=426 a=hat,b=wye,someothername=423 a=pan,b=hat,someothername=417 a=eks,b=hat,someothername=417 a=eks,b=eks,someothername=413 a=pan,b=zee,someothername=413 a=zee,b=hat,someothername=409 a=eks,b=wye,someothername=407 a=zee,b=zee,someothername=403 a=pan,b=wye,someothername=395 a=wye,b=pan,someothername=392 a=zee,b=eks,someothername=391 a=zee,b=pan,someothername=389 a=hat,b=eks,someothername=389 a=wye,b=eks,someothername=386 a=hat,b=zee,someothername=385 a=wye,b=zee,someothername=385 a=hat,b=hat,someothername=381 a=wye,b=wye,someothername=377 a=eks,b=pan,someothername=371 a=hat,b=pan,someothername=363 a=eks,b=zee,someothername=357
$ mlr count-distinct -n -f a,b data/medium count=25
count-similar
$ mlr count-similar --help
Usage: mlr count-similar [options]
Ingests all records, then emits each record augmented by a count of
the number of other records having the same group-by field values.
Options:
-g {d,e,f} Group-by-field names for counts.
-o {name} Field name for output count. Default "count".
$ mlr --opprint head -n 20 data/medium a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 wye pan 5 0.5732889198020006 0.8636244699032729 zee pan 6 0.5271261600918548 0.49322128674835697 eks zee 7 0.6117840605678454 0.1878849191181694 zee wye 8 0.5985540091064224 0.976181385699006 hat wye 9 0.03144187646093577 0.7495507603507059 pan wye 10 0.5026260055412137 0.9526183602969864 pan pan 11 0.7930488423451967 0.6505816637259333 zee pan 12 0.3676141320555616 0.23614420670296965 eks pan 13 0.4915175580479536 0.7709126592971468 eks zee 14 0.5207382318405251 0.34141681118811673 eks pan 15 0.07155556372719507 0.3596137145616235 pan pan 16 0.5736853980681922 0.7554169353781729 zee eks 17 0.29081949506712723 0.054478717073354166 hat zee 18 0.05727869223575699 0.13343527626645157 zee pan 19 0.43144132839222604 0.8442204830496998 eks wye 20 0.38245149780530685 0.4730652428100751
$ mlr --opprint head -n 20 then count-similar -g a data/medium a b i x y count pan pan 1 0.3467901443380824 0.7268028627434533 4 pan wye 10 0.5026260055412137 0.9526183602969864 4 pan pan 11 0.7930488423451967 0.6505816637259333 4 pan pan 16 0.5736853980681922 0.7554169353781729 4 eks pan 2 0.7586799647899636 0.5221511083334797 7 eks wye 4 0.38139939387114097 0.13418874328430463 7 eks zee 7 0.6117840605678454 0.1878849191181694 7 eks pan 13 0.4915175580479536 0.7709126592971468 7 eks zee 14 0.5207382318405251 0.34141681118811673 7 eks pan 15 0.07155556372719507 0.3596137145616235 7 eks wye 20 0.38245149780530685 0.4730652428100751 7 wye wye 3 0.20460330576630303 0.33831852551664776 2 wye pan 5 0.5732889198020006 0.8636244699032729 2 zee pan 6 0.5271261600918548 0.49322128674835697 5 zee wye 8 0.5985540091064224 0.976181385699006 5 zee pan 12 0.3676141320555616 0.23614420670296965 5 zee eks 17 0.29081949506712723 0.054478717073354166 5 zee pan 19 0.43144132839222604 0.8442204830496998 5 hat wye 9 0.03144187646093577 0.7495507603507059 2 hat zee 18 0.05727869223575699 0.13343527626645157 2
$ mlr --opprint head -n 20 then count-similar -g a then sort -f a data/medium a b i x y count eks pan 2 0.7586799647899636 0.5221511083334797 7 eks wye 4 0.38139939387114097 0.13418874328430463 7 eks zee 7 0.6117840605678454 0.1878849191181694 7 eks pan 13 0.4915175580479536 0.7709126592971468 7 eks zee 14 0.5207382318405251 0.34141681118811673 7 eks pan 15 0.07155556372719507 0.3596137145616235 7 eks wye 20 0.38245149780530685 0.4730652428100751 7 hat wye 9 0.03144187646093577 0.7495507603507059 2 hat zee 18 0.05727869223575699 0.13343527626645157 2 pan pan 1 0.3467901443380824 0.7268028627434533 4 pan wye 10 0.5026260055412137 0.9526183602969864 4 pan pan 11 0.7930488423451967 0.6505816637259333 4 pan pan 16 0.5736853980681922 0.7554169353781729 4 wye wye 3 0.20460330576630303 0.33831852551664776 2 wye pan 5 0.5732889198020006 0.8636244699032729 2 zee pan 6 0.5271261600918548 0.49322128674835697 5 zee wye 8 0.5985540091064224 0.976181385699006 5 zee pan 12 0.3676141320555616 0.23614420670296965 5 zee eks 17 0.29081949506712723 0.054478717073354166 5 zee pan 19 0.43144132839222604 0.8442204830496998 5
cut
$ mlr cut --help
Usage: mlr cut [options]
Passes through input records with specified fields included/excluded.
-f {a,b,c} Field names to include for cut.
-o Retain fields in the order specified here in the argument list.
Default is to retain them in the order found in the input data.
-x|--complement Exclude, rather than include, field names specified by -f.
-r Treat field names as regular expressions. "ab", "a.*b" will
match any field name containing the substring "ab" or matching
"a.*b", respectively; anchors of the form "^ab$", "^a.*b$" may
be used. The -o flag is ignored when -r is present.
Examples:
mlr cut -f hostname,status
mlr cut -x -f hostname,status
mlr cut -r -f '^status$,sda[0-9]'
mlr cut -r -f '^status$,"sda[0-9]"'
mlr cut -r -f '^status$,"sda[0-9]"i' (this is case-insensitive)
$ mlr --opprint cat data/small a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 wye pan 5 0.5732889198020006 0.8636244699032729 |
$ mlr --opprint cut -f y,x,i data/small i x y 1 0.3467901443380824 0.7268028627434533 2 0.7586799647899636 0.5221511083334797 3 0.20460330576630303 0.33831852551664776 4 0.38139939387114097 0.13418874328430463 5 0.5732889198020006 0.8636244699032729 |
$ echo 'a=1,b=2,c=3' | mlr cut -f b,c,a a=1,b=2,c=3 |
$ echo 'a=1,b=2,c=3' | mlr cut -o -f b,c,a b=2,c=3,a=1 |
decimate
$ mlr decimate --help
Usage: mlr decimate [options]
-n {count} Decimation factor; default 10
-b Decimate by printing first of every n.
-e Decimate by printing last of every n (default).
-g {a,b,c} Optional group-by-field names for decimate counts
Passes through one of every n records, optionally by category.
fill-down
$ mlr fill-down --help
Usage: mlr fill-down [options]
-f {a,b,c} Field names for fill-down
-a|--only-if-absent Field names for fill-down
If a given record has a missing value for a given field, fill that from
the corresponding value from a previous record, if any.
By default, a 'missing' field either is absent, or has the empty-string value.
With -a, a field is 'missing' only if it is absent.
$ cat data/fill-down.csv a,b,c 1,,3 4,5,6 7,,9
$ mlr --csv fill-down -f b data/fill-down.csv a,b,c 1,,3 4,5,6 7,5,9
$ mlr --csv fill-down -a -f b data/fill-down.csv a,b,c 1,,3 4,5,6 7,,9
filter
$ mlr filter --help
Usage: mlr filter [options] {expression}
Prints records for which {expression} evaluates to true.
If there are multiple semicolon-delimited expressions, all of them are
evaluated and the last one is used as the filter criterion.
Conversion options:
-S: Keeps field values as strings with no type inference to int or float.
-F: Keeps field values as strings or floats with no inference to int.
All field values are type-inferred to int/float/string unless this behavior is
suppressed with -S or -F.
Output/formatting options:
--oflatsep {string}: Separator to use when flattening multi-level @-variables
to output records for emit. Default ":".
--jknquoteint: For dump output (JSON-formatted), do not quote map keys if non-string.
--jvquoteall: For dump output (JSON-formatted), quote map values even if non-string.
Any of the output-format command-line flags (see mlr -h). Example: using
mlr --icsv --opprint ... then put --ojson 'tee > "mytap-".$a.".dat", $*' then ...
the input is CSV, the output is pretty-print tabular, but the tee-file output
is written in JSON format.
--no-fflush: for emit, tee, print, and dump, don't call fflush() after every
record.
Expression-specification options:
-f {filename}: the DSL expression is taken from the specified file rather
than from the command line. Outer single quotes wrapping the expression
should not be placed in the file. If -f is specified more than once,
all input files specified using -f are concatenated to produce the expression.
(For example, you can define functions in one file and call them from another.)
-e {expression}: You can use this after -f to add an expression. Example use
case: define functions/subroutines in a file you specify with -f, then call
them with an expression you specify with -e.
(If you mix -e and -f then the expressions are evaluated in the order encountered.
Since the expression pieces are simply concatenated, please be sure to use intervening
semicolons to separate expressions.)
Tracing options:
-v: Prints the expressions's AST (abstract syntax tree), which gives
full transparency on the precedence and associativity rules of
Miller's grammar, to stdout.
-a: Prints a low-level stack-allocation trace to stdout.
-t: Prints a low-level parser trace to stderr.
-T: Prints a every statement to stderr as it is executed.
Other options:
-x: Prints records for which {expression} evaluates to false.
Please use a dollar sign for field names and double-quotes for string
literals. If field names have special characters such as "." then you might
use braces, e.g. '${field.name}'. Miller built-in variables are
NF NR FNR FILENUM FILENAME M_PI M_E, and ENV["namegoeshere"] to access environment
variables. The environment-variable name may be an expression, e.g. a field
value.
Use # to comment to end of line.
Examples:
mlr filter 'log10($count) > 4.0'
mlr filter 'FNR == 2 (second record in each file)'
mlr filter 'urand() < 0.001' (subsampling)
mlr filter '$color != "blue" && $value > 4.2'
mlr filter '($x<.5 && $y<.5) || ($x>.5 && $y>.5)'
mlr filter '($name =~ "^sys.*east$") || ($name =~ "^dev.[0-9]+"i)'
mlr filter '$ab = $a+$b; $cd = $c+$d; $ab != $cd'
mlr filter '
NR == 1 ||
#NR == 2 ||
NR == 3
'
Please see http://johnkerl.org/miller/doc/reference.html for more information
including function list. Or "mlr -f". Please also also "mlr grep" which is
useful when you don't yet know which field name(s) you're looking for.
Features which filter shares with put
Please see Expression language for filter and put for more information about the expression language for mlr filter.fraction
$ mlr fraction --help
Usage: mlr fraction [options]
For each record's value in specified fields, computes the ratio of that
value to the sum of values in that field over all input records.
E.g. with input records x=1 x=2 x=3 and x=4, emits output records
x=1,x_fraction=0.1 x=2,x_fraction=0.2 x=3,x_fraction=0.3 and x=4,x_fraction=0.4
Note: this is internally a two-pass algorithm: on the first pass it retains
input records and accumulates sums; on the second pass it computes quotients
and emits output records. This means it produces no output until all input is read.
Options:
-f {a,b,c} Field name(s) for fraction calculation
-g {d,e,f} Optional group-by-field name(s) for fraction counts
-p Produce percents [0..100], not fractions [0..1]. Output field names
end with "_percent" rather than "_fraction"
-c Produce cumulative distributions, i.e. running sums: each output
value folds in the sum of the previous for the specified group
E.g. with input records x=1 x=2 x=3 and x=4, emits output records
x=1,x_cumulative_fraction=0.1 x=2,x_cumulative_fraction=0.3
x=3,x_cumulative_fraction=0.6 and x=4,x_cumulative_fraction=1.0
u=female,v=red,n=2458 u=female,v=green,n=192 u=female,v=blue,n=337 u=female,v=purple,n=468 u=female,v=yellow,n=3 u=female,v=orange,n=17 u=male,v=red,n=143 u=male,v=green,n=227 u=male,v=blue,n=2034 u=male,v=purple,n=12 u=male,v=yellow,n=1192 u=male,v=orange,n=448
$ mlr --opprint fraction -f n data/fraction-example.csv u v n n_fraction female red 2458 0.326384 female green 192 0.025495 female blue 337 0.044748 female purple 468 0.062143 female yellow 3 0.000398 female orange 17 0.002257 male red 143 0.018988 male green 227 0.030142 male blue 2034 0.270084 male purple 12 0.001593 male yellow 1192 0.158279 male orange 448 0.059487
$ mlr --opprint fraction -f n -g u data/fraction-example.csv u v n n_fraction female red 2458 0.707338 female green 192 0.055252 female blue 337 0.096978 female purple 468 0.134676 female yellow 3 0.000863 female orange 17 0.004892 male red 143 0.035256 male green 227 0.055966 male blue 2034 0.501479 male purple 12 0.002959 male yellow 1192 0.293886 male orange 448 0.110454 |
$ mlr --opprint fraction -f n -g v data/fraction-example.csv u v n n_fraction female red 2458 0.945021 female green 192 0.458234 female blue 337 0.142134 female purple 468 0.975000 female yellow 3 0.002510 female orange 17 0.036559 male red 143 0.054979 male green 227 0.541766 male blue 2034 0.857866 male purple 12 0.025000 male yellow 1192 0.997490 male orange 448 0.963441 |
$ mlr --opprint fraction -f n -p data/fraction-example.csv u v n n_percent female red 2458 32.638428 female green 192 2.549462 female blue 337 4.474837 female purple 468 6.214314 female yellow 3 0.039835 female orange 17 0.225734 male red 143 1.898818 male green 227 3.014208 male blue 2034 27.008365 male purple 12 0.159341 male yellow 1192 15.827911 male orange 448 5.948745
$ mlr --opprint fraction -f n -p -c data/fraction-example.csv u v n n_cumulative_percent female red 2458 32.638428 female green 192 35.187890 female blue 337 39.662727 female purple 468 45.877042 female yellow 3 45.916877 female orange 17 46.142611 male red 143 48.041429 male green 227 51.055637 male blue 2034 78.064002 male purple 12 78.223344 male yellow 1192 94.051255 male orange 448 100
$ mlr --opprint fraction -f n -g u -p -c data/fraction-example.csv u v n n_cumulative_percent female red 2458 70.733813 female green 192 76.258993 female blue 337 85.956835 female purple 468 99.424460 female yellow 3 99.510791 female orange 17 100 male red 143 3.525641 male green 227 9.122288 male blue 2034 59.270217 male purple 12 59.566075 male yellow 1192 88.954635 male orange 448 100
grep
$ mlr grep -h
Usage: mlr grep [options] {regular expression}
Passes through records which match {regex}.
Options:
-i Use case-insensitive search.
-v Invert: pass through records which do not match the regex.
Note that "mlr filter" is more powerful, but requires you to know field names.
By contrast, "mlr grep" allows you to regex-match the entire record. It does
this by formatting each record in memory as DKVP, using command-line-specified
ORS/OFS/OPS, and matching the resulting line against the regex specified
here. In particular, the regex is not applied to the input stream: if you
have CSV with header line "x,y,z" and data line "1,2,3" then the regex will
be matched, not against either of these lines, but against the DKVP line
"x=1,y=2,z=3". Furthermore, not all the options to system grep are supported,
and this command is intended to be merely a keystroke-saver. To get all the
features of system grep, you can do
"mlr --odkvp ... | grep ... | mlr --idkvp ..."
group-by
$ mlr group-by --help
Usage: mlr group-by {comma-separated field names}
Outputs records in batches having identical values at specified field names.
$ mlr --opprint group-by a data/small a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 eks wye 4 0.38139939387114097 0.13418874328430463 wye wye 3 0.20460330576630303 0.33831852551664776 wye pan 5 0.5732889198020006 0.8636244699032729 |
$ mlr --opprint sort -f a data/small a b i x y eks pan 2 0.7586799647899636 0.5221511083334797 eks wye 4 0.38139939387114097 0.13418874328430463 pan pan 1 0.3467901443380824 0.7268028627434533 wye wye 3 0.20460330576630303 0.33831852551664776 wye pan 5 0.5732889198020006 0.8636244699032729 |
group-like
$ mlr group-like --help Usage: mlr group-like Outputs records in batches having identical field names.
$ mlr cat data/het.dkvp resource=/path/to/file,loadsec=0.45,ok=true record_count=100,resource=/path/to/file resource=/path/to/second/file,loadsec=0.32,ok=true record_count=150,resource=/path/to/second/file resource=/some/other/path,loadsec=0.97,ok=false |
$ mlr --opprint group-like data/het.dkvp resource loadsec ok /path/to/file 0.45 true /path/to/second/file 0.32 true /some/other/path 0.97 false record_count resource 100 /path/to/file 150 /path/to/second/file |
having-fields
$ mlr having-fields --help
Usage: mlr having-fields [options]
Conditionally passes through records depending on each record's field names.
Options:
--at-least {comma-separated names}
--which-are {comma-separated names}
--at-most {comma-separated names}
--all-matching {regular expression}
--any-matching {regular expression}
--none-matching {regular expression}
Examples:
mlr having-fields --which-are amount,status,owner
mlr having-fields --any-matching 'sda[0-9]'
mlr having-fields --any-matching '"sda[0-9]"'
mlr having-fields --any-matching '"sda[0-9]"i' (this is case-insensitive)
$ mlr cat data/het.dkvp resource=/path/to/file,loadsec=0.45,ok=true record_count=100,resource=/path/to/file resource=/path/to/second/file,loadsec=0.32,ok=true record_count=150,resource=/path/to/second/file resource=/some/other/path,loadsec=0.97,ok=false |
$ mlr having-fields --at-least resource data/het.dkvp resource=/path/to/file,loadsec=0.45,ok=true record_count=100,resource=/path/to/file resource=/path/to/second/file,loadsec=0.32,ok=true record_count=150,resource=/path/to/second/file resource=/some/other/path,loadsec=0.97,ok=false |
$ mlr having-fields --which-are resource,ok,loadsec data/het.dkvp resource=/path/to/file,loadsec=0.45,ok=true resource=/path/to/second/file,loadsec=0.32,ok=true resource=/some/other/path,loadsec=0.97,ok=false |
head
$ mlr head --help
Usage: mlr head [options]
-n {count} Head count to print; default 10
-g {a,b,c} Optional group-by-field names for head counts
Passes through the first n records, optionally by category.
Without -g, ceases consuming more input (i.e. is fast) when n
records have been read.
$ mlr --opprint head -n 4 data/medium a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 |
$ mlr --opprint head -n 1 -g b data/medium a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 wye wye 3 0.20460330576630303 0.33831852551664776 eks zee 7 0.6117840605678454 0.1878849191181694 zee eks 17 0.29081949506712723 0.054478717073354166 wye hat 24 0.7286126830627567 0.19441962592638418 |
histogram
$ mlr histogram --help
Usage: mlr histogram [options]
-f {a,b,c} Value-field names for histogram counts
--lo {lo} Histogram low value
--hi {hi} Histogram high value
--nbins {n} Number of histogram bins
--auto Automatically computes limits, ignoring --lo and --hi.
Holds all values in memory before producing any output.
-o {prefix} Prefix for output field name. Default: no prefix.
Just a histogram. Input values < lo or > hi are not counted.
$ mlr --opprint put '$x2=$x**2;$x3=$x2*$x' then histogram -f x,x2,x3 --lo 0 --hi 1 --nbins 10 data/medium bin_lo bin_hi x_count x2_count x3_count 0.000000 0.100000 1072 3231 4661 0.100000 0.200000 938 1254 1184 0.200000 0.300000 1037 988 845 0.300000 0.400000 988 832 676 0.400000 0.500000 950 774 576 0.500000 0.600000 1002 692 476 0.600000 0.700000 1007 591 438 0.700000 0.800000 1007 560 420 0.800000 0.900000 986 571 383 0.900000 1.000000 1013 507 341
$ mlr --opprint put '$x2=$x**2;$x3=$x2*$x' then histogram -f x,x2,x3 --lo 0 --hi 1 --nbins 10 -o my_ data/medium my_bin_lo my_bin_hi my_x_count my_x2_count my_x3_count 0.000000 0.100000 1072 3231 4661 0.100000 0.200000 938 1254 1184 0.200000 0.300000 1037 988 845 0.300000 0.400000 988 832 676 0.400000 0.500000 950 774 576 0.500000 0.600000 1002 692 476 0.600000 0.700000 1007 591 438 0.700000 0.800000 1007 560 420 0.800000 0.900000 986 571 383 0.900000 1.000000 1013 507 341
join
$ mlr join --help
Usage: mlr join [options]
Joins records from specified left file name with records from all file names
at the end of the Miller argument list.
Functionality is essentially the same as the system "join" command, but for
record streams.
Options:
-f {left file name}
-j {a,b,c} Comma-separated join-field names for output
-l {a,b,c} Comma-separated join-field names for left input file;
defaults to -j values if omitted.
-r {a,b,c} Comma-separated join-field names for right input file(s);
defaults to -j values if omitted.
--lp {text} Additional prefix for non-join output field names from
the left file
--rp {text} Additional prefix for non-join output field names from
the right file(s)
--np Do not emit paired records
--ul Emit unpaired records from the left file
--ur Emit unpaired records from the right file(s)
-s|--sorted-input Require sorted input: records must be sorted
lexically by their join-field names, else not all records will
be paired. The only likely use case for this is with a left
file which is too big to fit into system memory otherwise.
-u Enable unsorted input. (This is the default even without -u.)
In this case, the entire left file will be loaded into memory.
--prepipe {command} As in main input options; see mlr --help for details.
If you wish to use a prepipe command for the main input as well
as here, it must be specified there as well as here.
File-format options default to those for the right file names on the Miller
argument list, but may be overridden for the left file as follows. Please see
the main "mlr --help" for more information on syntax for these arguments.
-i {one of csv,dkvp,nidx,pprint,xtab}
--irs {record-separator character}
--ifs {field-separator character}
--ips {pair-separator character}
--repifs
--repips
--mmap
--no-mmap
Please use "mlr --usage-separator-options" for information on specifying separators.
Please see http://johnkerl.org/miller/doc/reference.html for more information
including examples.
$ mlr --icsvlite --opprint cat data/join-left-example.csv id name 100 alice 200 bob 300 carol 400 david 500 edgar |
$ mlr --icsvlite --opprint cat data/join-right-example.csv status idcode present 400 present 100 missing 200 present 100 present 200 missing 100 missing 200 present 300 missing 600 present 400 present 400 present 300 present 100 missing 400 present 200 present 200 present 200 present 200 present 400 present 300 |
$ mlr --icsvlite --opprint join -u -j id -r idcode -f data/join-left-example.csv data/join-right-example.csv id name status 400 david present 100 alice present 200 bob missing 100 alice present 200 bob present 100 alice missing 200 bob missing 300 carol present 400 david present 400 david present 300 carol present 100 alice present 400 david missing 200 bob present 200 bob present 200 bob present 200 bob present 400 david present 300 carol present |
$ mlr --icsvlite --opprint sort -f idcode then join -j id -r idcode -f data/join-left-example.csv data/join-right-example.csv id name status 100 alice present 100 alice present 100 alice missing 100 alice present 200 bob missing 200 bob present 200 bob missing 200 bob present 200 bob present 200 bob present 200 bob present 300 carol present 300 carol present 300 carol present 400 david present 400 david present 400 david present 400 david missing 400 david present |
$ mlr --icsvlite --opprint join --np --ul --ur -u -j id -r idcode -f data/join-left-example.csv data/join-right-example.csv status idcode missing 600 id name 500 edgar |
$ mlr --csvlite --opprint cat data/self-join.csv data/self-join.csv a b c 1 2 3 1 4 5 1 2 3 1 4 5 |
$ mlr --csvlite --opprint join -j a --lp left_ --rp right_ -f data/self-join.csv data/self-join.csv a left_b left_c right_b right_c 1 2 3 2 3 1 4 5 2 3 1 2 3 4 5 1 4 5 4 5 |
$ mlr --csvlite --opprint join -j "" --lp left_ --rp right_ -f data/self-join.csv data/self-join.csv left_a left_b left_c right_a right_b right_c 1 2 3 1 2 3 1 4 5 1 2 3 1 2 3 1 4 5 1 4 5 1 4 5 |
label
$ mlr label --help
Usage: mlr label {new1,new2,new3,...}
Given n comma-separated names, renames the first n fields of each record to
have the respective name. (Fields past the nth are left with their original
names.) Particularly useful with --inidx or --implicit-csv-header, to give
useful names to otherwise integer-indexed fields.
Examples:
"echo 'a b c d' | mlr --inidx --odkvp cat" gives "1=a,2=b,3=c,4=d"
"echo 'a b c d' | mlr --inidx --odkvp label s,t" gives "s=a,t=b,3=c,4=d"
% grep -v '^#' /etc/passwd | mlr --nidx --fs : --opprint label name,password,uid,gid,gecos,home_dir,shell | head name password uid gid gecos home_dir shell nobody * -2 -2 Unprivileged User /var/empty /usr/bin/false root * 0 0 System Administrator /var/root /bin/sh daemon * 1 1 System Services /var/root /usr/bin/false _uucp * 4 4 Unix to Unix Copy Protocol /var/spool/uucp /usr/sbin/uucico _taskgated * 13 13 Task Gate Daemon /var/empty /usr/bin/false _networkd * 24 24 Network Services /var/networkd /usr/bin/false _installassistant * 25 25 Install Assistant /var/empty /usr/bin/false _lp * 26 26 Printing Services /var/spool/cups /usr/bin/false _postfix * 27 27 Postfix Mail Server /var/spool/postfix /usr/bin/false
$ cat data/headerless.csv John,23,present Fred,34,present Alice,56,missing Carol,45,present
$ mlr --csv --implicit-csv-header cat data/headerless.csv 1,2,3 John,23,present Fred,34,present Alice,56,missing Carol,45,present
$ mlr --csv --implicit-csv-header label name,age,status data/headerless.csv name,age,status John,23,present Fred,34,present Alice,56,missing Carol,45,present
$ mlr --icsv --implicit-csv-header --opprint label name,age,status data/headerless.csv name age status John 23 present Fred 34 present Alice 56 missing Carol 45 present
least-frequent
$ mlr least-frequent -h
Usage: mlr least-frequent [options]
Shows the least frequently occurring distinct values for specified field names.
The first entry is the statistical anti-mode; the remaining are runners-up.
Options:
-f {one or more comma-separated field names}. Required flag.
-n {count}. Optional flag defaulting to 10.
-b Suppress counts; show only field values.
-o {name} Field name for output count. Default "count".
See also "mlr most-frequent".
$ mlr --opprint --from data/colored-shapes.dkvp least-frequent -f shape -n 5 shape count circle 2591 triangle 3372 square 4115
$ mlr --opprint --from data/colored-shapes.dkvp least-frequent -f shape,color -n 5 shape color count circle orange 68 triangle orange 107 square orange 128 circle green 287 circle purple 289
$ mlr --opprint --from data/colored-shapes.dkvp least-frequent -f shape,color -n 5 -o someothername shape color someothername circle orange 68 triangle orange 107 square orange 128 circle green 287 circle purple 289
$ mlr --opprint --from data/colored-shapes.dkvp least-frequent -f shape,color -n 5 -b shape color circle orange triangle orange square orange circle green circle purple
merge-fields
$ mlr merge-fields --help
Usage: mlr merge-fields [options]
Computes univariate statistics for each input record, accumulated across
specified fields.
Options:
-a {sum,count,...} Names of accumulators. One or more of:
count Count instances of fields
mode Find most-frequently-occurring values for fields; first-found wins tie
antimode Find least-frequently-occurring values for fields; first-found wins tie
sum Compute sums of specified fields
mean Compute averages (sample means) of specified fields
stddev Compute sample standard deviation of specified fields
var Compute sample variance of specified fields
meaneb Estimate error bars for averages (assuming no sample autocorrelation)
skewness Compute sample skewness of specified fields
kurtosis Compute sample kurtosis of specified fields
min Compute minimum values of specified fields
max Compute maximum values of specified fields
-f {a,b,c} Value-field names on which to compute statistics. Requires -o.
-r {a,b,c} Regular expressions for value-field names on which to compute
statistics. Requires -o.
-c {a,b,c} Substrings for collapse mode. All fields which have the same names
after removing substrings will be accumulated together. Please see
examples below.
-i Use interpolated percentiles, like R's type=7; default like type=1.
Not sensical for string-valued fields.
-o {name} Output field basename for -f/-r.
-k Keep the input fields which contributed to the output statistics;
the default is to omit them.
-F Computes integerable things (e.g. count) in floating point.
String-valued data make sense unless arithmetic on them is required,
e.g. for sum, mean, interpolated percentiles, etc. In case of mixed data,
numbers are less than strings.
Example input data: "a_in_x=1,a_out_x=2,b_in_y=4,b_out_x=8".
Example: mlr merge-fields -a sum,count -f a_in_x,a_out_x -o foo
produces "b_in_y=4,b_out_x=8,foo_sum=3,foo_count=2" since "a_in_x,a_out_x" are
summed over.
Example: mlr merge-fields -a sum,count -r in_,out_ -o bar
produces "bar_sum=15,bar_count=4" since all four fields are summed over.
Example: mlr merge-fields -a sum,count -c in_,out_
produces "a_x_sum=3,a_x_count=2,b_y_sum=4,b_y_count=1,b_x_sum=8,b_x_count=1"
since "a_in_x" and "a_out_x" both collapse to "a_x", "b_in_y" collapses to
"b_y", and "b_out_x" collapses to "b_x".
$ mlr --csvlite --opprint cat data/inout.csv a_in a_out b_in b_out 436 490 446 195 526 320 963 780 220 888 705 831
$ mlr --csvlite --opprint merge-fields -a min,max,sum -c _in,_out data/inout.csv a_min a_max a_sum b_min b_max b_sum 436 490 926 195 446 641 320 526 846 780 963 1743 220 888 1108 705 831 1536
$ mlr --csvlite --opprint merge-fields -k -a sum -c _in,_out data/inout.csv a_in a_out b_in b_out a_sum b_sum 436 490 446 195 926 641 526 320 963 780 846 1743 220 888 705 831 1108 1536
most-frequent
$ mlr most-frequent -h
Usage: mlr most-frequent [options]
Shows the most frequently occurring distinct values for specified field names.
The first entry is the statistical mode; the remaining are runners-up.
Options:
-f {one or more comma-separated field names}. Required flag.
-n {count}. Optional flag defaulting to 10.
-b Suppress counts; show only field values.
-o {name} Field name for output count. Default "count".
See also "mlr least-frequent".
$ mlr --opprint --from data/colored-shapes.dkvp most-frequent -f shape -n 5 shape count square 4115 triangle 3372 circle 2591
$ mlr --opprint --from data/colored-shapes.dkvp most-frequent -f shape,color -n 5 shape color count square red 1874 triangle red 1560 circle red 1207 square yellow 589 square blue 589
$ mlr --opprint --from data/colored-shapes.dkvp most-frequent -f shape,color -n 5 -o someothername shape color someothername square red 1874 triangle red 1560 circle red 1207 square yellow 589 square blue 589
$ mlr --opprint --from data/colored-shapes.dkvp most-frequent -f shape,color -n 5 -b shape color square red triangle red circle red square yellow square blue
nest
$ mlr nest -h
Usage: mlr nest [options]
Explodes specified field values into separate fields/records, or reverses this.
Options:
--explode,--implode One is required.
--values,--pairs One is required.
--across-records,--across-fields One is required.
-f {field name} Required.
--nested-fs {string} Defaults to ";". Field separator for nested values.
--nested-ps {string} Defaults to ":". Pair separator for nested key-value pairs.
--evar {string} Shorthand for --explode --values ---across-records --nested-fs {string}
Please use "mlr --usage-separator-options" for information on specifying separators.
Examples:
mlr nest --explode --values --across-records -f x
with input record "x=a;b;c,y=d" produces output records
"x=a,y=d"
"x=b,y=d"
"x=c,y=d"
Use --implode to do the reverse.
mlr nest --explode --values --across-fields -f x
with input record "x=a;b;c,y=d" produces output records
"x_1=a,x_2=b,x_3=c,y=d"
Use --implode to do the reverse.
mlr nest --explode --pairs --across-records -f x
with input record "x=a:1;b:2;c:3,y=d" produces output records
"a=1,y=d"
"b=2,y=d"
"c=3,y=d"
mlr nest --explode --pairs --across-fields -f x
with input record "x=a:1;b:2;c:3,y=d" produces output records
"a=1,b=2,c=3,y=d"
Notes:
* With --pairs, --implode doesn't make sense since the original field name has
been lost.
* The combination "--implode --values --across-records" is non-streaming:
no output records are produced until all input records have been read. In
particular, this means it won't work in tail -f contexts. But all other flag
combinations result in streaming (tail -f friendly) data processing.
* It's up to you to ensure that the nested-fs is distinct from your data's IFS:
e.g. by default the former is semicolon and the latter is comma.
See also mlr reshape.
nothing
$ mlr nothing -h Usage: mlr nothing [options] Drops all input records. Useful for testing, or after tee/print/etc. have produced other output.
put
$ mlr put --help
Usage: mlr put [options] {expression}
Adds/updates specified field(s). Expressions are semicolon-separated and must
either be assignments, or evaluate to boolean. Booleans with following
statements in curly braces control whether those statements are executed;
booleans without following curly braces do nothing except side effects (e.g.
regex-captures into \1, \2, etc.).
Conversion options:
-S: Keeps field values as strings with no type inference to int or float.
-F: Keeps field values as strings or floats with no inference to int.
All field values are type-inferred to int/float/string unless this behavior is
suppressed with -S or -F.
Output/formatting options:
--oflatsep {string}: Separator to use when flattening multi-level @-variables
to output records for emit. Default ":".
--jknquoteint: For dump output (JSON-formatted), do not quote map keys if non-string.
--jvquoteall: For dump output (JSON-formatted), quote map values even if non-string.
Any of the output-format command-line flags (see mlr -h). Example: using
mlr --icsv --opprint ... then put --ojson 'tee > "mytap-".$a.".dat", $*' then ...
the input is CSV, the output is pretty-print tabular, but the tee-file output
is written in JSON format.
--no-fflush: for emit, tee, print, and dump, don't call fflush() after every
record.
Expression-specification options:
-f {filename}: the DSL expression is taken from the specified file rather
than from the command line. Outer single quotes wrapping the expression
should not be placed in the file. If -f is specified more than once,
all input files specified using -f are concatenated to produce the expression.
(For example, you can define functions in one file and call them from another.)
-e {expression}: You can use this after -f to add an expression. Example use
case: define functions/subroutines in a file you specify with -f, then call
them with an expression you specify with -e.
(If you mix -e and -f then the expressions are evaluated in the order encountered.
Since the expression pieces are simply concatenated, please be sure to use intervening
semicolons to separate expressions.)
Tracing options:
-v: Prints the expressions's AST (abstract syntax tree), which gives
full transparency on the precedence and associativity rules of
Miller's grammar, to stdout.
-a: Prints a low-level stack-allocation trace to stdout.
-t: Prints a low-level parser trace to stderr.
-T: Prints a every statement to stderr as it is executed.
Other options:
-q: Does not include the modified record in the output stream. Useful for when
all desired output is in begin and/or end blocks.
Please use a dollar sign for field names and double-quotes for string
literals. If field names have special characters such as "." then you might
use braces, e.g. '${field.name}'. Miller built-in variables are
NF NR FNR FILENUM FILENAME M_PI M_E, and ENV["namegoeshere"] to access environment
variables. The environment-variable name may be an expression, e.g. a field
value.
Use # to comment to end of line.
Examples:
mlr put '$y = log10($x); $z = sqrt($y)'
mlr put '$x>0.0 { $y=log10($x); $z=sqrt($y) }' # does {...} only if $x > 0.0
mlr put '$x>0.0; $y=log10($x); $z=sqrt($y)' # does all three statements
mlr put '$a =~ "([a-z]+)_([0-9]+); $b = "left_\1"; $c = "right_\2"'
mlr put '$a =~ "([a-z]+)_([0-9]+) { $b = "left_\1"; $c = "right_\2" }'
mlr put '$filename = FILENAME'
mlr put '$colored_shape = $color . "_" . $shape'
mlr put '$y = cos($theta); $z = atan2($y, $x)'
mlr put '$name = sub($name, "http.*com"i, "")'
mlr put -q '@sum += $x; end {emit @sum}'
mlr put -q '@sum[$a] += $x; end {emit @sum, "a"}'
mlr put -q '@sum[$a][$b] += $x; end {emit @sum, "a", "b"}'
mlr put -q '@min=min(@min,$x);@max=max(@max,$x); end{emitf @min, @max}'
mlr put -q 'is_null(@xmax) || $x > @xmax {@xmax=$x; @recmax=$*}; end {emit @recmax}'
mlr put '
$x = 1;
#$y = 2;
$z = 3
'
Please see also 'mlr -k' for examples using redirected output.
Please see http://johnkerl.org/miller/doc/reference.html for more information
including function list. Or "mlr -f".
Please see in particular:
http://www.johnkerl.org/miller/doc/reference.html#put
Features which put shares with filter
Please see Expression language for filter and put for more information about the expression language for mlr put.regularize
$ mlr regularize --help Usage: mlr regularize For records seen earlier in the data stream with same field names in a different order, outputs them with field names in the previously encountered order. Example: input records a=1,c=2,b=3, then e=4,d=5, then c=7,a=6,b=8 output as a=1,c=2,b=3, then e=4,d=5, then a=6,c=7,b=8
rename
$ mlr rename --help
Usage: mlr rename [options] {old1,new1,old2,new2,...}
Renames specified fields.
Options:
-r Treat old field names as regular expressions. "ab", "a.*b"
will match any field name containing the substring "ab" or
matching "a.*b", respectively; anchors of the form "^ab$",
"^a.*b$" may be used. New field names may be plain strings,
or may contain capture groups of the form "\1" through
"\9". Wrapping the regex in double quotes is optional, but
is required if you wish to follow it with 'i' to indicate
case-insensitivity.
-g Do global replacement within each field name rather than
first-match replacement.
Examples:
mlr rename old_name,new_name'
mlr rename old_name_1,new_name_1,old_name_2,new_name_2'
mlr rename -r 'Date_[0-9]+,Date,' Rename all such fields to be "Date"
mlr rename -r '"Date_[0-9]+",Date' Same
mlr rename -r 'Date_([0-9]+).*,\1' Rename all such fields to be of the form 20151015
mlr rename -r '"name"i,Name' Rename "name", "Name", "NAME", etc. to "Name"
$ mlr --opprint cat data/small a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 wye pan 5 0.5732889198020006 0.8636244699032729 |
$ mlr --opprint rename i,INDEX,b,COLUMN2 data/small a COLUMN2 INDEX x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 wye pan 5 0.5732889198020006 0.8636244699032729 |
$ sed 's/y/COLUMN5/g' data/small a=pan,b=pan,i=1,x=0.3467901443380824,COLUMN5=0.7268028627434533 a=eks,b=pan,i=2,x=0.7586799647899636,COLUMN5=0.5221511083334797 a=wCOLUMN5e,b=wCOLUMN5e,i=3,x=0.20460330576630303,COLUMN5=0.33831852551664776 a=eks,b=wCOLUMN5e,i=4,x=0.38139939387114097,COLUMN5=0.13418874328430463 a=wCOLUMN5e,b=pan,i=5,x=0.5732889198020006,COLUMN5=0.8636244699032729 |
$ mlr rename y,COLUMN5 data/small a=pan,b=pan,i=1,x=0.3467901443380824,COLUMN5=0.7268028627434533 a=eks,b=pan,i=2,x=0.7586799647899636,COLUMN5=0.5221511083334797 a=wye,b=wye,i=3,x=0.20460330576630303,COLUMN5=0.33831852551664776 a=eks,b=wye,i=4,x=0.38139939387114097,COLUMN5=0.13418874328430463 a=wye,b=pan,i=5,x=0.5732889198020006,COLUMN5=0.8636244699032729 |
reorder
$ mlr reorder --help
Usage: mlr reorder [options]
-f {a,b,c} Field names to reorder.
-e Put specified field names at record end: default is to put
them at record start.
Examples:
mlr reorder -f a,b sends input record "d=4,b=2,a=1,c=3" to "a=1,b=2,d=4,c=3".
mlr reorder -e -f a,b sends input record "d=4,b=2,a=1,c=3" to "d=4,c=3,a=1,b=2".
$ mlr --opprint cat data/small a b i x y pan pan 1 0.3467901443380824 0.7268028627434533 eks pan 2 0.7586799647899636 0.5221511083334797 wye wye 3 0.20460330576630303 0.33831852551664776 eks wye 4 0.38139939387114097 0.13418874328430463 wye pan 5 0.5732889198020006 0.8636244699032729 | |
$ mlr --opprint reorder -f i,b data/small i b a x y 1 pan pan 0.3467901443380824 0.7268028627434533 2 pan eks 0.7586799647899636 0.5221511083334797 3 wye wye 0.20460330576630303 0.33831852551664776 4 wye eks 0.38139939387114097 0.13418874328430463 5 pan wye 0.5732889198020006 0.8636244699032729 |
$ mlr --opprint reorder -e -f i,b data/small a x y i b pan 0.3467901443380824 0.7268028627434533 1 pan eks 0.7586799647899636 0.5221511083334797 2 pan wye 0.20460330576630303 0.33831852551664776 3 wye eks 0.38139939387114097 0.13418874328430463 4 wye wye 0.5732889198020006 0.8636244699032729 5 pan |
repeat
$ mlr repeat --help
Usage: mlr repeat [options]
Copies input records to output records multiple times.
Options must be exactly one of the following:
-n {repeat count} Repeat each input record this many times.
-f {field name} Same, but take the repeat count from the specified
field name of each input record.
Example:
echo x=0 | mlr repeat -n 4 then put '$x=urand()'
produces:
x=0.488189
x=0.484973
x=0.704983
x=0.147311
Example:
echo a=1,b=2,c=3 | mlr repeat -f b
produces:
a=1,b=2,c=3
a=1,b=2,c=3
Example:
echo a=1,b=2,c=3 | mlr repeat -f c
produces:
a=1,b=2,c=3
a=1,b=2,c=3
a=1,b=2,c=3
This is useful in at least two ways: one, as a data-generator as in the above example using urand(); two, for reconstructing individual samples from data which has been count-aggregated:
$ cat data/repeat-example.dat color=blue,count=5 color=red,count=4 color=green,count=3
$ mlr repeat -f count then cut -x -f count data/repeat-example.dat color=blue color=blue color=blue color=blue color=blue color=red color=red color=red color=red color=green color=green color=green
After expansion with repeat, such data can then be sent on to stats1 -a mode, or (if the data are numeric) to stats1 -a p10,p50,p90, etc.
reshape
$ mlr reshape --help
Usage: mlr reshape [options]
Wide-to-long options:
-i {input field names} -o {key-field name,value-field name}
-r {input field regexes} -o {key-field name,value-field name}
These pivot/reshape the input data such that the input fields are removed
and separate records are emitted for each key/value pair.
Note: this works with tail -f and produces output records for each input
record seen.
Long-to-wide options:
-s {key-field name,value-field name}
These pivot/reshape the input data to undo the wide-to-long operation.
Note: this does not work with tail -f; it produces output records only after
all input records have been read.
Examples:
Input file "wide.txt":
time X Y
2009-01-01 0.65473572 2.4520609
2009-01-02 -0.89248112 0.2154713
2009-01-03 0.98012375 1.3179287
mlr --pprint reshape -i X,Y -o item,value wide.txt
time item value
2009-01-01 X 0.65473572
2009-01-01 Y 2.4520609
2009-01-02 X -0.89248112
2009-01-02 Y 0.2154713
2009-01-03 X 0.98012375
2009-01-03 Y 1.3179287
mlr --pprint reshape -r '[A-Z]' -o item,value wide.txt
time item value
2009-01-01 X 0.65473572
2009-01-01 Y 2.4520609
2009-01-02 X -0.89248112
2009-01-02 Y 0.2154713
2009-01-03 X 0.98012375
2009-01-03 Y 1.3179287
Input file "long.txt":
time item value
2009-01-01 X 0.65473572
2009-01-01 Y 2.4520609
2009-01-02 X -0.89248112
2009-01-02 Y 0.2154713
2009-01-03 X 0.98012375
2009-01-03 Y 1.3179287
mlr --pprint reshape -s item,value long.txt
time X Y
2009-01-01 0.65473572 2.4520609
2009-01-02 -0.89248112 0.2154713
2009-01-03 0.98012375 1.3179287
See also mlr nest.
sample
$ mlr sample --help
Usage: mlr sample [options]
Reservoir sampling (subsampling without replacement), optionally by category.
-k {count} Required: number of records to output, total, or by group if using -g.
-g {a,b,c} Optional: group-by-field names for samples.
See also mlr bootstrap and mlr shuffle.
$ mlr --opprint sample -k 4 data/colored-shapes.dkvp color shape flag i u v w x purple triangle 0 90122 0.9986871176198068 0.3037738877233719 0.5154934457238382 5.365962021016529 red circle 0 3139 0.04835898233323954 -0.03964684310055758 0.5263660881848111 5.3758779366493625 orange triangle 0 67847 0.36746306902109926 0.5161574810505635 0.5176199566173642 3.1748088656576567 yellow square 1 33576 0.3098376725521097 0.8525628505287842 0.49774122460981685 4.494754378604669 $ mlr --opprint sample -k 4 data/colored-shapes.dkvp color shape flag i u v w x blue square 1 16783 0.09974385090654347 0.7243899920872646 0.5353718443278438 4.431057737383438 orange square 1 93291 0.5944176543007182 0.17744449786454086 0.49262281749172077 3.1548117990710653 yellow square 1 54436 0.5268161165014636 0.8785588662666121 0.5058773791931063 7.019185838783636 yellow square 1 55491 0.0025440267883102274 0.05474106287787284 0.5102729153751984 3.526301273728043 $ mlr --opprint sample -k 2 -g color data/colored-shapes.dkvp color shape flag i u v w x yellow triangle 1 11 0.6321695890307647 0.9887207810889004 0.4364983936735774 5.7981881667050565 yellow square 1 917 0.8547010348386344 0.7356782810796262 0.4531511689924275 5.774541777078352 red circle 1 4000 0.05490416175132373 0.07392337815122155 0.49416101516594396 5.355725080701707 red square 0 87506 0.6357719216821314 0.6970867759393995 0.4940826462055272 6.351579417310387 purple triangle 0 14898 0.7800986870203719 0.23998073813992293 0.5014775988383656 3.141006771777843 purple triangle 0 151 0.032614487569017414 0.7346633365041219 0.7812143304483805 2.6831992610568047 green triangle 1 126 0.1513010528347546 0.40346767294704544 0.051213231883952326 5.955109300797182 green circle 0 17635 0.029856606049114442 0.4724542934246524 0.49529606749929744 5.239153910272168 blue circle 1 1020 0.414263129226617 0.8304946402876182 0.13151094520189244 4.397873687920433 blue triangle 0 220 0.441773289968473 0.44597731903759075 0.6329360666849821 4.3064608776550894 orange square 0 1885 0.8079311983747106 0.8685956833908394 0.3116410800256374 4.390864584500387 orange triangle 0 1533 0.32904497195507487 0.23168161807490417 0.8722623057355134 5.164071635714438 $ mlr --opprint sample -k 2 -g color then sort -f color data/colored-shapes.dkvp color shape flag i u v w x blue circle 0 215 0.7803586969333292 0.33146680638888126 0.04289047852629113 5.725365736377487 blue circle 1 3616 0.8548431579124808 0.4989623130006362 0.3339426415875795 3.696785877560498 green square 0 356 0.7674272008085286 0.341578843118008 0.4570224877870851 4.830320062215299 green square 0 152 0.6684429446914862 0.016056003736548696 0.4656148241291592 5.434588759225423 orange triangle 0 587 0.5175826237797857 0.08989091493635304 0.9011709461770973 4.265854207755811 orange triangle 0 1533 0.32904497195507487 0.23168161807490417 0.8722623057355134 5.164071635714438 purple triangle 0 14192 0.5196327866973567 0.7860928603468063 0.4964368415453642 4.899167143824484 purple triangle 0 65 0.6842806710360729 0.5823723856331258 0.8014053396013747 5.805148213865135 red square 1 2431 0.38378504852300466 0.11445015005595527 0.49355539228753786 5.146756570128739 red triangle 0 57097 0.43763430414406546 0.3355450325004481 0.5322349637512487 4.144267240289442 yellow triangle 1 11 0.6321695890307647 0.9887207810889004 0.4364983936735774 5.7981881667050565 yellow square 1 158 0.41527900739142165 0.7118027080775757 0.4200799665161291 5.33279067554884
sec2gmt
$ mlr sec2gmt -h
Usage: mlr sec2gmt [options] {comma-separated list of field names}
Replaces a numeric field representing seconds since the epoch with the
corresponding GMT timestamp; leaves non-numbers as-is. This is nothing
more than a keystroke-saver for the sec2gmt function:
mlr sec2gmt time1,time2
is the same as
mlr put '$time1=sec2gmt($time1);$time2=sec2gmt($time2)'
Options:
-1 through -9: format the seconds using 1..9 decimal places, respectively.
sec2gmtdate
$ mlr sec2gmtdate -h
Usage: mlr sec2gmtdate {comma-separated list of field names}
Replaces a numeric field representing seconds since the epoch with the
corresponding GMT year-month-day timestamp; leaves non-numbers as-is.
This is nothing more than a keystroke-saver for the sec2gmtdate function:
mlr sec2gmtdate time1,time2
is the same as
mlr put '$time1=sec2gmtdate($time1);$time2=sec2gmtdate($time2)'
seqgen
$ mlr seqgen -h
Usage: mlr seqgen [options]
Produces a sequence of counters. Discards the input record stream. Produces
output as specified by the following options:
-f {name} Field name for counters; default "i".
--start {number} Inclusive start value; default "1".
--stop {number} Inclusive stop value; default "100".
--step {number} Step value; default "1".
Start, stop, and/or step may be floating-point. Output is integer if start,
stop, and step are all integers. Step may be negative. It may not be zero
unless start == stop.
$ mlr seqgen --stop 10 i=1 i=2 i=3 i=4 i=5 i=6 i=7 i=8 i=9 i=10
$ mlr seqgen --start 20 --stop 40 --step 4 i=20 i=24 i=28 i=32 i=36 i=40
$ mlr seqgen --start 40 --stop 20 --step -4 i=40 i=36 i=32 i=28 i=24 i=20
shuffle
$ mlr shuffle -h
Usage: mlr shuffle {no options}
Outputs records randomly permuted. No output records are produced until
all input records are read.
See also mlr bootstrap and mlr sample.
sort
$ mlr sort --help
Usage: mlr sort {flags}
Flags:
-f {comma-separated field names} Lexical ascending
-n {comma-separated field names} Numerical ascending; nulls sort last
-nf {comma-separated field names} Numerical ascending; nulls sort last
-r {comma-separated field names} Lexical descending
-nr {comma-separated field names} Numerical descending; nulls sort first
Sorts records primarily by the first specified field, secondarily by the second
field, and so on. (Any records not having all specified sort keys will appear
at the end of the output, in the order they were encountered, regardless of the
specified sort order.) The sort is stable: records that compare equal will sort
in the order they were encountered in the input record stream.
Example:
mlr sort -f a,b -nr x,y,z
which is the same as:
mlr sort -f a -f b -nr x -nr y -nr z
$ mlr --opprint sort -f a -nr x data/small a b i x y eks pan 2 0.7586799647899636 0.5221511083334797 eks wye 4 0.38139939387114097 0.13418874328430463 pan pan 1 0.3467901443380824 0.7268028627434533 wye pan 5 0.5732889198020006 0.8636244699032729 wye wye 3 0.20460330576630303 0.33831852551664776
$ head -n 10 data/multicountdown.dat upsec=0.002,color=green,count=1203 upsec=0.083,color=red,count=3817 upsec=0.188,color=red,count=3801 upsec=0.395,color=blue,count=2697 upsec=0.526,color=purple,count=953 upsec=0.671,color=blue,count=2684 upsec=0.899,color=purple,count=926 upsec=0.912,color=red,count=3798 upsec=1.093,color=blue,count=2662 upsec=1.327,color=purple,count=917
$ head -n 20 data/multicountdown.dat | mlr --opprint sort -f color upsec color count 0.395 blue 2697 0.671 blue 2684 1.093 blue 2662 2.064 blue 2659 2.2880000000000003 blue 2647 0.002 green 1203 1.407 green 1187 1.448 green 1177 2.313 green 1161 0.526 purple 953 0.899 purple 926 1.327 purple 917 1.703 purple 908 0.083 red 3817 0.188 red 3801 0.912 red 3798 1.416 red 3788 1.587 red 3782 1.601 red 3755 1.832 red 3717
$ mlr sort -n x data/sort-missing.dkvp x=1 x=2 x=4 a=3
$ mlr sort -nr x data/sort-missing.dkvp x=4 x=2 x=1 a=3
stats1
$ mlr stats1 --help
Usage: mlr stats1 [options]
Computes univariate statistics for one or more given fields, accumulated across
the input record stream.
Options:
-a {sum,count,...} Names of accumulators: p10 p25.2 p50 p98 p100 etc. and/or
one or more of:
count Count instances of fields
mode Find most-frequently-occurring values for fields; first-found wins tie
antimode Find least-frequently-occurring values for fields; first-found wins tie
sum Compute sums of specified fields
mean Compute averages (sample means) of specified fields
stddev Compute sample standard deviation of specified fields
var Compute sample variance of specified fields
meaneb Estimate error bars for averages (assuming no sample autocorrelation)
skewness Compute sample skewness of specified fields
kurtosis Compute sample kurtosis of specified fields
min Compute minimum values of specified fields
max Compute maximum values of specified fields
-f {a,b,c} Value-field names on which to compute statistics
--fr {regex} Regex for value-field names on which to compute statistics
(compute statsitics on values in all field names matching regex)
--fx {regex} Inverted regex for value-field names on which to compute statistics
(compute statsitics on values in all field names not matching regex)
-g {d,e,f} Optional group-by-field names
--gr {regex} Regex for optional group-by-field names
(group by values in field names matching regex)
--gx {regex} Inverted regex for optional group-by-field names
(group by values in field names not matching regex)
--grfx {regex} Shorthand for --gr {regex} --fx {that same regex}
-i Use interpolated percentiles, like R's type=7; default like type=1.
Not sensical for string-valued fields.
-s Print iterative stats. Useful in tail -f contexts (in which
case please avoid pprint-format output since end of input
stream will never be seen).
-F Computes integerable things (e.g. count) in floating point.
Example: mlr stats1 -a min,p10,p50,p90,max -f value -g size,shape
Example: mlr stats1 -a count,mode -f size
Example: mlr stats1 -a count,mode -f size -g shape
Example: mlr stats1 -a count,mode --fr '^[a-h].*$' -gr '^k.*$'
This computes count and mode statistics on all field names beginning
with a through h, grouped by all field names starting with k.
Notes:
* p50 and median are synonymous.
* min and max output the same results as p0 and p100, respectively, but use
less memory.
* String-valued data make sense unless arithmetic on them is required,
e.g. for sum, mean, interpolated percentiles, etc. In case of mixed data,
numbers are less than strings.
* count and mode allow text input; the rest require numeric input.
In particular, 1 and 1.0 are distinct text for count and mode.
* When there are mode ties, the first-encountered datum wins.
$ mlr --oxtab stats1 -a count,sum,min,p10,p50,mean,p90,max -f x,y data/medium x_count 10000 x_sum 4986.019682 x_min 0.000045 x_p10 0.093322 x_p50 0.501159 x_mean 0.498602 x_p90 0.900794 x_max 0.999953 y_count 10000 y_sum 5062.057445 y_min 0.000088 y_p10 0.102132 y_p50 0.506021 y_mean 0.506206 y_p90 0.905366 y_max 0.999965 |
$ mlr --opprint stats1 -a mean -f x,y -g b then sort -f b data/medium b x_mean y_mean eks 0.506361 0.510293 hat 0.487899 0.513118 pan 0.497304 0.499599 wye 0.497593 0.504596 zee 0.504242 0.502997 |
$ mlr --opprint stats1 -a p50,p99 -f u,v -g color then put '$ur=$u_p99/$u_p50;$vr=$v_p99/$v_p50' data/colored-shapes.dkvp color u_p50 u_p99 v_p50 v_p99 ur vr yellow 0.501019 0.989046 0.520630 0.987034 1.974069 1.895845 red 0.485038 0.990054 0.492586 0.994444 2.041189 2.018823 purple 0.501319 0.988893 0.504571 0.988287 1.972582 1.958668 green 0.502015 0.990764 0.505359 0.990175 1.973574 1.959350 blue 0.525226 0.992655 0.485170 0.993873 1.889958 2.048505 orange 0.483548 0.993635 0.480913 0.989102 2.054884 2.056717 |
$ mlr --opprint count-distinct -f shape then sort -nr count data/colored-shapes.dkvp shape count square 4115 triangle 3372 circle 2591 |
$ mlr --opprint stats1 -a mode -f color -g shape data/colored-shapes.dkvp shape color_mode triangle red square red circle red |
stats2
$ mlr stats2 --help
Usage: mlr stats2 [options]
Computes bivariate statistics for one or more given field-name pairs,
accumulated across the input record stream.
-a {linreg-ols,corr,...} Names of accumulators: one or more of:
linreg-pca Linear regression using principal component analysis
linreg-ols Linear regression using ordinary least squares
r2 Quality metric for linreg-ols (linreg-pca emits its own)
logireg Logistic regression
corr Sample correlation
cov Sample covariance
covx Sample-covariance matrix
-f {a,b,c,d} Value-field name-pairs on which to compute statistics.
There must be an even number of names.
-g {e,f,g} Optional group-by-field names.
-v Print additional output for linreg-pca.
-s Print iterative stats. Useful in tail -f contexts (in which
case please avoid pprint-format output since end of input
stream will never be seen).
--fit Rather than printing regression parameters, applies them to
the input data to compute new fit fields. All input records are
held in memory until end of input stream. Has effect only for
linreg-ols, linreg-pca, and logireg.
Only one of -s or --fit may be used.
Example: mlr stats2 -a linreg-pca -f x,y
Example: mlr stats2 -a linreg-ols,r2 -f x,y -g size,shape
Example: mlr stats2 -a corr -f x,y
$ mlr --oxtab put '$x2=$x*$x; $xy=$x*$y; $y2=$y**2' then stats2 -a cov,corr -f x,y,y,y,x2,xy,x2,y2 data/medium x_y_cov 0.000043 x_y_corr 0.000504 y_y_cov 0.084611 y_y_corr 1.000000 x2_xy_cov 0.041884 x2_xy_corr 0.630174 x2_y2_cov -0.000310 x2_y2_corr -0.003425 |
$ mlr --opprint put '$x2=$x*$x; $xy=$x*$y; $y2=$y**2' then stats2 -a linreg-ols,r2 -f x,y,y,y,xy,y2 -g a data/medium a x_y_ols_m x_y_ols_b x_y_ols_n x_y_r2 y_y_ols_m y_y_ols_b y_y_ols_n y_y_r2 xy_y2_ols_m xy_y2_ols_b xy_y2_ols_n xy_y2_r2 pan 0.017026 0.500403 2081 0.000287 1.000000 0.000000 2081 1.000000 0.878132 0.119082 2081 0.417498 eks 0.040780 0.481402 1965 0.001646 1.000000 0.000000 1965 1.000000 0.897873 0.107341 1965 0.455632 wye -0.039153 0.525510 1966 0.001505 1.000000 0.000000 1966 1.000000 0.853832 0.126745 1966 0.389917 zee 0.002781 0.504307 2047 0.000008 1.000000 0.000000 2047 1.000000 0.852444 0.124017 2047 0.393566 hat -0.018621 0.517901 1941 0.000352 1.000000 0.000000 1941 1.000000 0.841230 0.135573 1941 0.368794 |
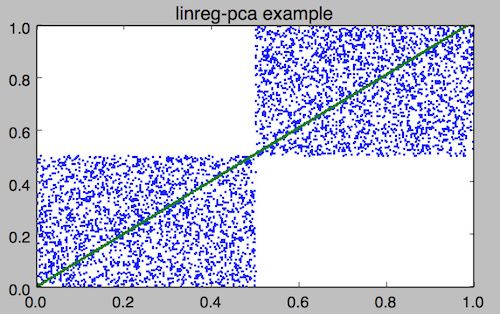
# Prepare input data: mlr filter '($x<.5 && $y<.5) || ($x>.5 && $y>.5)' data/medium > data/medium-squares # Do a linear regression and examine coefficients: mlr --ofs newline stats2 -a linreg-pca -f x,y data/medium-squares x_y_pca_m=1.014419 x_y_pca_b=0.000308 x_y_pca_quality=0.861354 # Option 1 to apply the regression coefficients and produce a linear fit: # Set x_y_pca_m and x_y_pca_b as shell variables: eval $(mlr --ofs newline stats2 -a linreg-pca -f x,y data/medium-squares) # In addition to x and y, make a new yfit which is the line fit, then plot # using your favorite tool: mlr --onidx put '$yfit='$x_y_pca_m'*$x+'$x_y_pca_b then cut -x -f a,b,i data/medium-squares \ | pgr -p -title 'linreg-pca example' -xmin 0 -xmax 1 -ymin 0 -ymax 1 # Option 2 to apply the regression coefficients and produce a linear fit: use --fit option mlr --onidx stats2 -a linreg-pca --fit -f x,y then cut -f a,b,i data/medium-squares \ | pgr -p -title 'linreg-pca example' -xmin 0 -xmax 1 -ymin 0 -ymax 1
$ head -n 10 data/multicountdown.dat upsec=0.002,color=green,count=1203 upsec=0.083,color=red,count=3817 upsec=0.188,color=red,count=3801 upsec=0.395,color=blue,count=2697 upsec=0.526,color=purple,count=953 upsec=0.671,color=blue,count=2684 upsec=0.899,color=purple,count=926 upsec=0.912,color=red,count=3798 upsec=1.093,color=blue,count=2662 upsec=1.327,color=purple,count=917
$ mlr --oxtab stats2 -a linreg-pca -f upsec,count -g color then put '$donesec = -$upsec_count_pca_b/$upsec_count_pca_m' data/multicountdown.dat color green upsec_count_pca_m -32.756917 upsec_count_pca_b 1213.722730 upsec_count_pca_n 24 upsec_count_pca_quality 0.999984 donesec 37.052410 color red upsec_count_pca_m -37.367646 upsec_count_pca_b 3810.133400 upsec_count_pca_n 30 upsec_count_pca_quality 0.999989 donesec 101.963431 color blue upsec_count_pca_m -29.231212 upsec_count_pca_b 2698.932820 upsec_count_pca_n 25 upsec_count_pca_quality 0.999959 donesec 92.330514 color purple upsec_count_pca_m -39.030097 upsec_count_pca_b 979.988341 upsec_count_pca_n 21 upsec_count_pca_quality 0.999991 donesec 25.108529
step
$ mlr step --help
Usage: mlr step [options]
Computes values dependent on the previous record, optionally grouped
by category.
Options:
-a {delta,rsum,...} Names of steppers: comma-separated, one or more of:
delta Compute differences in field(s) between successive records
shift Include value(s) in field(s) from previous record, if any
from-first Compute differences in field(s) from first record
ratio Compute ratios in field(s) between successive records
rsum Compute running sums of field(s) between successive records
counter Count instances of field(s) between successive records
ewma Exponentially weighted moving average over successive records
-f {a,b,c} Value-field names on which to compute statistics
-g {d,e,f} Optional group-by-field names
-F Computes integerable things (e.g. counter) in floating point.
-d {x,y,z} Weights for ewma. 1 means current sample gets all weight (no
smoothing), near under under 1 is light smoothing, near over 0 is
heavy smoothing. Multiple weights may be specified, e.g.
"mlr step -a ewma -f sys_load -d 0.01,0.1,0.9". Default if omitted
is "-d 0.5".
-o {a,b,c} Custom suffixes for EWMA output fields. If omitted, these default to
the -d values. If supplied, the number of -o values must be the same
as the number of -d values.
Examples:
mlr step -a rsum -f request_size
mlr step -a delta -f request_size -g hostname
mlr step -a ewma -d 0.1,0.9 -f x,y
mlr step -a ewma -d 0.1,0.9 -o smooth,rough -f x,y
mlr step -a ewma -d 0.1,0.9 -o smooth,rough -f x,y -g group_name
Please see http://johnkerl.org/miller/doc/reference.html#filter or
https://en.wikipedia.org/wiki/Moving_average#Exponential_moving_average
for more information on EWMA.
$ mlr --opprint step -a shift,delta,rsum,counter -f x data/medium | head -15 a b i x y x_shift x_delta x_rsum x_counter pan pan 1 0.3467901443380824 0.7268028627434533 - 0 0.346790 1 eks pan 2 0.7586799647899636 0.5221511083334797 0.3467901443380824 0.411890 1.105470 2 wye wye 3 0.20460330576630303 0.33831852551664776 0.7586799647899636 -0.554077 1.310073 3 eks wye 4 0.38139939387114097 0.13418874328430463 0.20460330576630303 0.176796 1.691473 4 wye pan 5 0.5732889198020006 0.8636244699032729 0.38139939387114097 0.191890 2.264762 5 zee pan 6 0.5271261600918548 0.49322128674835697 0.5732889198020006 -0.046163 2.791888 6 eks zee 7 0.6117840605678454 0.1878849191181694 0.5271261600918548 0.084658 3.403672 7 zee wye 8 0.5985540091064224 0.976181385699006 0.6117840605678454 -0.013230 4.002226 8 hat wye 9 0.03144187646093577 0.7495507603507059 0.5985540091064224 -0.567112 4.033668 9 pan wye 10 0.5026260055412137 0.9526183602969864 0.03144187646093577 0.471184 4.536294 10 pan pan 11 0.7930488423451967 0.6505816637259333 0.5026260055412137 0.290423 5.329343 11 zee pan 12 0.3676141320555616 0.23614420670296965 0.7930488423451967 -0.425435 5.696957 12 eks pan 13 0.4915175580479536 0.7709126592971468 0.3676141320555616 0.123903 6.188474 13 eks zee 14 0.5207382318405251 0.34141681118811673 0.4915175580479536 0.029221 6.709213 14 |
$ mlr --opprint step -a shift,delta,rsum,counter -f x -g a data/medium | head -15 a b i x y x_shift x_delta x_rsum x_counter pan pan 1 0.3467901443380824 0.7268028627434533 - 0 0.346790 1 eks pan 2 0.7586799647899636 0.5221511083334797 - 0 0.758680 1 wye wye 3 0.20460330576630303 0.33831852551664776 - 0 0.204603 1 eks wye 4 0.38139939387114097 0.13418874328430463 0.7586799647899636 -0.377281 1.140079 2 wye pan 5 0.5732889198020006 0.8636244699032729 0.20460330576630303 0.368686 0.777892 2 zee pan 6 0.5271261600918548 0.49322128674835697 - 0 0.527126 1 eks zee 7 0.6117840605678454 0.1878849191181694 0.38139939387114097 0.230385 1.751863 3 zee wye 8 0.5985540091064224 0.976181385699006 0.5271261600918548 0.071428 1.125680 2 hat wye 9 0.03144187646093577 0.7495507603507059 - 0 0.031442 1 pan wye 10 0.5026260055412137 0.9526183602969864 0.3467901443380824 0.155836 0.849416 2 pan pan 11 0.7930488423451967 0.6505816637259333 0.5026260055412137 0.290423 1.642465 3 zee pan 12 0.3676141320555616 0.23614420670296965 0.5985540091064224 -0.230940 1.493294 3 eks pan 13 0.4915175580479536 0.7709126592971468 0.6117840605678454 -0.120267 2.243381 4 eks zee 14 0.5207382318405251 0.34141681118811673 0.4915175580479536 0.029221 2.764119 5 |
$ mlr --opprint step -a ewma -f x -d 0.1,0.9 ../doc/data/medium | head -15 a b i x y x_ewma_0.1 x_ewma_0.9 pan pan 1 0.3467901443380824 0.7268028627434533 0.346790 0.346790 eks pan 2 0.7586799647899636 0.5221511083334797 0.387979 0.717491 wye wye 3 0.20460330576630303 0.33831852551664776 0.369642 0.255892 eks wye 4 0.38139939387114097 0.13418874328430463 0.370817 0.368849 wye pan 5 0.5732889198020006 0.8636244699032729 0.391064 0.552845 zee pan 6 0.5271261600918548 0.49322128674835697 0.404671 0.529698 eks zee 7 0.6117840605678454 0.1878849191181694 0.425382 0.603575 zee wye 8 0.5985540091064224 0.976181385699006 0.442699 0.599056 hat wye 9 0.03144187646093577 0.7495507603507059 0.401573 0.088203 pan wye 10 0.5026260055412137 0.9526183602969864 0.411679 0.461184 pan pan 11 0.7930488423451967 0.6505816637259333 0.449816 0.759862 zee pan 12 0.3676141320555616 0.23614420670296965 0.441596 0.406839 eks pan 13 0.4915175580479536 0.7709126592971468 0.446588 0.483050 eks zee 14 0.5207382318405251 0.34141681118811673 0.454003 0.516969 |
$ mlr --opprint step -a ewma -f x -d 0.1,0.9 -o smooth,rough ../doc/data/medium | head -15 a b i x y x_ewma_smooth x_ewma_rough pan pan 1 0.3467901443380824 0.7268028627434533 0.346790 0.346790 eks pan 2 0.7586799647899636 0.5221511083334797 0.387979 0.717491 wye wye 3 0.20460330576630303 0.33831852551664776 0.369642 0.255892 eks wye 4 0.38139939387114097 0.13418874328430463 0.370817 0.368849 wye pan 5 0.5732889198020006 0.8636244699032729 0.391064 0.552845 zee pan 6 0.5271261600918548 0.49322128674835697 0.404671 0.529698 eks zee 7 0.6117840605678454 0.1878849191181694 0.425382 0.603575 zee wye 8 0.5985540091064224 0.976181385699006 0.442699 0.599056 hat wye 9 0.03144187646093577 0.7495507603507059 0.401573 0.088203 pan wye 10 0.5026260055412137 0.9526183602969864 0.411679 0.461184 pan pan 11 0.7930488423451967 0.6505816637259333 0.449816 0.759862 zee pan 12 0.3676141320555616 0.23614420670296965 0.441596 0.406839 eks pan 13 0.4915175580479536 0.7709126592971468 0.446588 0.483050 eks zee 14 0.5207382318405251 0.34141681118811673 0.454003 0.516969 |
$ each 10 uptime | mlr -p step -a delta -f 11 ... 20:08 up 36 days, 10:38, 5 users, load averages: 1.42 1.62 1.73 0.000000 20:08 up 36 days, 10:38, 5 users, load averages: 1.55 1.64 1.74 0.020000 20:08 up 36 days, 10:38, 7 users, load averages: 1.58 1.65 1.74 0.010000 20:08 up 36 days, 10:38, 9 users, load averages: 1.78 1.69 1.76 0.040000 20:08 up 36 days, 10:39, 9 users, load averages: 2.12 1.76 1.78 0.070000 20:08 up 36 days, 10:39, 9 users, load averages: 2.51 1.85 1.81 0.090000 20:08 up 36 days, 10:39, 8 users, load averages: 2.79 1.92 1.83 0.070000 20:08 up 36 days, 10:39, 4 users, load averages: 2.64 1.90 1.83 -0.020000
tac
$ mlr tac --help Usage: mlr tac Prints records in reverse order from the order in which they were encountered.
$ mlr --icsv --opprint cat data/a.csv a b c 1 2 3 4 5 6 |
$ mlr --icsv --opprint cat data/b.csv a b c 7 8 9 |
$ mlr --icsv --opprint tac data/a.csv data/b.csv a b c 7 8 9 4 5 6 1 2 3 |
$ mlr --icsv --opprint put '$filename=FILENAME' then tac data/a.csv data/b.csv a b c filename 7 8 9 data/b.csv 4 5 6 data/a.csv 1 2 3 data/a.csv |
tail
$ mlr tail --help
Usage: mlr tail [options]
-n {count} Tail count to print; default 10
-g {a,b,c} Optional group-by-field names for tail counts
Passes through the last n records, optionally by category.
$ mlr --opprint tail -n 4 data/colored-shapes.dkvp color shape flag i u v w x blue square 1 99974 0.6189062525431605 0.2637962404841453 0.5311465405784674 6.210738209085753 blue triangle 0 99976 0.008110504040268474 0.8267274952432482 0.4732962944898885 6.146956761817328 yellow triangle 0 99990 0.3839424618160777 0.55952913620132 0.5113763011485609 4.307973891915119 yellow circle 1 99994 0.764950884927175 0.25284227383991364 0.49969878539567425 5.013809741826425 |
$ mlr --opprint tail -n 1 -g shape data/colored-shapes.dkvp color shape flag i u v w x yellow triangle 0 99990 0.3839424618160777 0.55952913620132 0.5113763011485609 4.307973891915119 blue square 1 99974 0.6189062525431605 0.2637962404841453 0.5311465405784674 6.210738209085753 yellow circle 1 99994 0.764950884927175 0.25284227383991364 0.49969878539567425 5.013809741826425 |
tee
$ mlr tee --help
Usage: mlr tee [options] {filename}
Passes through input records (like mlr cat) but also writes to specified output
file, using output-format flags from the command line (e.g. --ocsv). See also
the "tee" keyword within mlr put, which allows data-dependent filenames.
Options:
-a: append to existing file, if any, rather than overwriting.
--no-fflush: don't call fflush() after every record.
Any of the output-format command-line flags (see mlr -h). Example: using
mlr --icsv --opprint put '...' then tee --ojson ./mytap.dat then stats1 ...
the input is CSV, the output is pretty-print tabular, but the tee-file output
is written in JSON format.
top
$ mlr top --help
Usage: mlr top [options]
-f {a,b,c} Value-field names for top counts.
-g {d,e,f} Optional group-by-field names for top counts.
-n {count} How many records to print per category; default 1.
-a Print all fields for top-value records; default is
to print only value and group-by fields. Requires a single
value-field name only.
--min Print top smallest values; default is top largest values.
-F Keep top values as floats even if they look like integers.
-o {name} Field name for output indices. Default "top_idx".
Prints the n records with smallest/largest values at specified fields,
optionally by category.
$ mlr --opprint top -n 4 -f x data/medium top_idx x_top 1 0.999953 2 0.999823 3 0.999733 4 0.999563 $ mlr --opprint top -n 4 -f x -o someothername data/medium someothername x_top 1 0.999953 2 0.999823 3 0.999733 4 0.999563 |
$ mlr --opprint top -n 2 -f x -g a then sort -f a data/medium a top_idx x_top eks 1 0.998811 eks 2 0.998534 hat 1 0.999953 hat 2 0.999733 pan 1 0.999403 pan 2 0.999044 wye 1 0.999823 wye 2 0.999264 zee 1 0.999490 zee 2 0.999438 |
uniq
$ mlr uniq --help
Usage: mlr uniq [options]
Prints distinct values for specified field names. With -c, same as
count-distinct. For uniq, -f is a synonym for -g.
Options:
-g {d,e,f} Group-by-field names for uniq counts.
-c Show repeat counts in addition to unique values.
-n Show only the number of distinct values.
-o {name} Field name for output count. Default "count".
-a Output each unique record only once. Incompatible with -g.
With -c, produces unique records, with repeat counts for each.
With -n, produces only one record which is the unique-record count.
With neither -c nor -n, produces unique records.
$ wc -l data/colored-shapes.dkvp 10078 data/colored-shapes.dkvp |
$ mlr uniq -g color,shape data/colored-shapes.dkvp color=yellow,shape=triangle color=red,shape=square color=red,shape=circle color=purple,shape=triangle color=yellow,shape=circle color=purple,shape=square color=yellow,shape=square color=red,shape=triangle color=green,shape=triangle color=green,shape=square color=blue,shape=circle color=blue,shape=triangle color=purple,shape=circle color=blue,shape=square color=green,shape=circle color=orange,shape=triangle color=orange,shape=square color=orange,shape=circle |
$ mlr --opprint uniq -g color,shape -c then sort -f color,shape data/colored-shapes.dkvp color shape count blue circle 384 blue square 589 blue triangle 497 green circle 287 green square 454 green triangle 368 orange circle 68 orange square 128 orange triangle 107 purple circle 289 purple square 481 purple triangle 372 red circle 1207 red square 1874 red triangle 1560 yellow circle 356 yellow square 589 yellow triangle 468 |
$ mlr --opprint uniq -g color,shape -c -o someothername then sort -nr someothername data/colored-shapes.dkvp color shape someothername red square 1874 red triangle 1560 red circle 1207 yellow square 589 blue square 589 blue triangle 497 purple square 481 yellow triangle 468 green square 454 blue circle 384 purple triangle 372 green triangle 368 yellow circle 356 purple circle 289 green circle 287 orange square 128 orange triangle 107 orange circle 68 |
$ mlr --opprint uniq -n -g color,shape data/colored-shapes.dkvp count 18 |
$ cat data/repeats.dkvp color=red,shape=square,flag=0 color=purple,shape=triangle,flag=0 color=yellow,shape=circle,flag=1 color=red,shape=circle,flag=1 color=red,shape=square,flag=0 color=yellow,shape=circle,flag=1 color=red,shape=square,flag=0 color=red,shape=square,flag=0 color=yellow,shape=circle,flag=1 color=red,shape=circle,flag=1 color=yellow,shape=circle,flag=1 color=yellow,shape=circle,flag=1 color=purple,shape=triangle,flag=0 color=yellow,shape=circle,flag=1 color=yellow,shape=circle,flag=1 color=red,shape=circle,flag=1 color=red,shape=square,flag=0 color=purple,shape=triangle,flag=0 color=yellow,shape=circle,flag=1 color=red,shape=square,flag=0 color=purple,shape=square,flag=0 color=red,shape=square,flag=0 color=red,shape=square,flag=1 color=red,shape=square,flag=0 color=red,shape=square,flag=0 color=purple,shape=triangle,flag=0 color=red,shape=square,flag=0 color=purple,shape=triangle,flag=0 color=red,shape=square,flag=0 color=red,shape=square,flag=0 color=purple,shape=square,flag=0 color=red,shape=square,flag=0 color=red,shape=square,flag=0 color=purple,shape=triangle,flag=0 color=yellow,shape=triangle,flag=1 color=purple,shape=square,flag=0 color=yellow,shape=circle,flag=1 color=purple,shape=triangle,flag=0 color=red,shape=circle,flag=1 color=purple,shape=triangle,flag=0 color=purple,shape=triangle,flag=0 color=red,shape=square,flag=0 color=red,shape=circle,flag=1 color=red,shape=square,flag=1 color=red,shape=square,flag=0 color=red,shape=circle,flag=1 color=purple,shape=square,flag=0 color=purple,shape=square,flag=0 color=red,shape=square,flag=1 color=purple,shape=triangle,flag=0 color=purple,shape=triangle,flag=0 color=purple,shape=square,flag=0 color=yellow,shape=circle,flag=1 color=red,shape=square,flag=0 color=yellow,shape=triangle,flag=1 color=yellow,shape=circle,flag=1 color=purple,shape=square,flag=0 | |
$ wc -l data/repeats.dkvp
57 data/repeats.dkvp
|
$ mlr --opprint uniq -a data/repeats.dkvp color shape flag red square 0 purple triangle 0 yellow circle 1 red circle 1 purple square 0 red square 1 yellow triangle 1 |
$ mlr --opprint uniq -a -n data/repeats.dkvp count 7 |
$ mlr --opprint uniq -a -c data/repeats.dkvp count color shape flag 17 red square 0 11 purple triangle 0 11 yellow circle 1 6 red circle 1 7 purple square 0 3 red square 1 2 yellow triangle 1 |
unsparsify
$ mlr unsparsify --help
Usage: mlr unsparsify [options]
Prints records with the union of field names over all input records.
For field names absent in a given record but present in others, fills in
a value. This verb retains all input before producing any output.
Options:
--fill-with {filler string} What to fill absent fields with. Defaults to
the empty string.
Example: if the input is two records, one being 'a=1,b=2' and the other
being 'b=3,c=4', then the output is the two records 'a=1,b=2,c=' and
'a=,b=3,c=4'.
$ cat data/sparse.json
{"a":1,"b":2,"v":3}
{"u":1,"b":2}
{"a":1,"v":2,"x":3}
{"v":1,"w":2}
$ mlr --json unsparsify data/sparse.json
{ "a": 1, "b": 2, "v": 3, "u": "", "x": "", "w": "" }
{ "a": "", "b": 2, "v": "", "u": 1, "x": "", "w": "" }
{ "a": 1, "b": "", "v": 2, "u": "", "x": 3, "w": "" }
{ "a": "", "b": "", "v": 1, "u": "", "x": "", "w": 2 }
$ mlr --ijson --opprint unsparsify data/sparse.json a b v u x w 1 2 3 - - - - 2 - 1 - - 1 - 2 - 3 - - - 1 - - 2
$ mlr --ijson --opprint unsparsify --fill-with missing data/sparse.json a b v u x w 1 2 3 missing missing missing missing 2 missing 1 missing missing 1 missing 2 missing 3 missing missing missing 1 missing missing 2